| Localtab |
|---|
| active | true |
|---|
| title | Data Access |
|---|
| Data Access
| Data Type | Download all or Query/Filter |
|---|
Click the Download button to save a ".tcia" manifest file to your computer, which you must open with the NBIA Data Retriever . Click the Search button to open our Data Portal, where you can browse the data collection and/or download a subset of its contents. | Download all or Query/Filter | License |
|---|
776 | Target Data (XLSX) 131 (KB) | | | Biopsy Data (XLSX) 4.25 (MB) | | | | | Biopsy Overlays (ZIP) 333 (MB) | | Click the Versions tab for more info about data releases.
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/Prostate-MRI-US-Biopsy-NBIA-manifest_v2_20231020.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=Prostate-MRI-US-Biopsy |
|---|
|
|
(Download requires the NBIA Data Retriever) | | Target Data (XLSX, 131 KB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/Target-Data_2019-12-05%20%282%29.xlsx?api=v2 |
|---|
|
|
| | | Biopsy Data (XLSX, 4.25 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/TCIA%20Biopsy%20Data_2020-07-14.xlsx?api=v2 |
|---|
|
|
| | |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYzNSIsInBhc3Njb2RlIjoiNzZkZDBhOWY2Y2YzZjQ1OGQ0ODZlOWE4OTRiOGExMjRjNmNkOGUwNyIsInBhY2thZ2VfaWQiOiI2MzUiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | | Biopsy Overlays (ZIP, 333 MB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYzMyIsInBhc3Njb2RlIjoiZjY3MzUxMGUyMGY5OGY3YzFhMDE4MDBmZWFjYTRiYWZiYjFjNDE4NCIsInBhY2thZ2VfaWQiOiI2MzMiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | |
Click the Versions tab for more info about data releases. | Nci_crdc additional resources |
|---|
|
| Localtab |
|---|
| title | Detailed Description |
|---|
| Detailed Description
Image Statistics | Radiology Image Statistics |
|---|
Modalities | MR, US | Number of Participants | 1,151 | Number of Studies | 2,799 | Number of Series | 4,819 | Number of Images | 102,397 | | Images Size (GB) | 79.4 | Instructions for Proper Use of the STL FilesThe 3D surfaces of this dataset are in stereolithography format (binary representation). They are segmentations representing anatomic regions within the corresponding ultrasound (US) or MRI images. Each file is named "Prostate-MRI-US-Biopsy-XXXX-SURFACETYPE-seriesUID-YYYY.STL" where "XXXX" is the anonymized patient number and "YYYY" is the series instance UID of corresponding DICOM images. "SURFACETYPE" can be one of two identifiers, summarized below. - If "SURFACETYPE" is "ProstateSurface," then the STL file is a segmentation of the prostate gland. These were contoured semi-automatically with Profuse (for MRI) or the Artemis fusion biopsy system (for US), both products of Eigen (Grass Valley, CA). Using these tools, the user manually traced the prostate on one or more image planes, and the remainder of gland was automatically segmented in 3D. The user was then able to adjust vertices of the 3D segmentation as necessary. These segmentations are generally of good quality, though their accuracy is user-dependent.
- If "SURFACETYPE" is "Target," then the STL file is a segmentation of a Region of Interest (ROI) suspicious for harboring prostate cancer. Zero, one, or more than one ROI may be present, and thus are designated "Target1," "Target2," etc. The ROIs corresponding to MRI were traced manually by a radiologist based on their interpretation of the patient's multiparametric MRI study. The ROIs corresponding to US images were acquired by registering the MRI prostate surface nonrigidly and semi-automatically to the US prostate surface. Therefore, the US ROIs are subject to misregistration, with average accuracy of 3-4 mm.
- Note that proximity to an ROI increases the likelihood of, but does not guarantee, prostate cancer. Also note that radiologists assigned each ROI a Likert-like suspicion score of 1-5, with 1 corresponding to very low suspicion of prostate cancer and 5 corresponding to very high suspicion of prostate cancer. These scores, which are roughly equivalent to the "PIRADS Version 2" scoring system, can be found in a spreadsheet included with the dataset.
The STL files can be visualized in 3D Slicer as follows: - Import the surface(s) either by drag-and-dropping or with File->Add Data
- Within the "Models" module, adjust Display->Opacity of the prostate surface in order to visualize any ROI(s) within
- To visualize surfaces in the context of corresponding MRI or US images, load the corresponding image series and then toggle the Slice Display->Visible radio button within the "models" module
- To visualize surfaces in the context of biopsy cores, import the corresponding 3D slicer file named "BiopsyVectors.mrml" by drag-and-dropping or with File->Add Data.
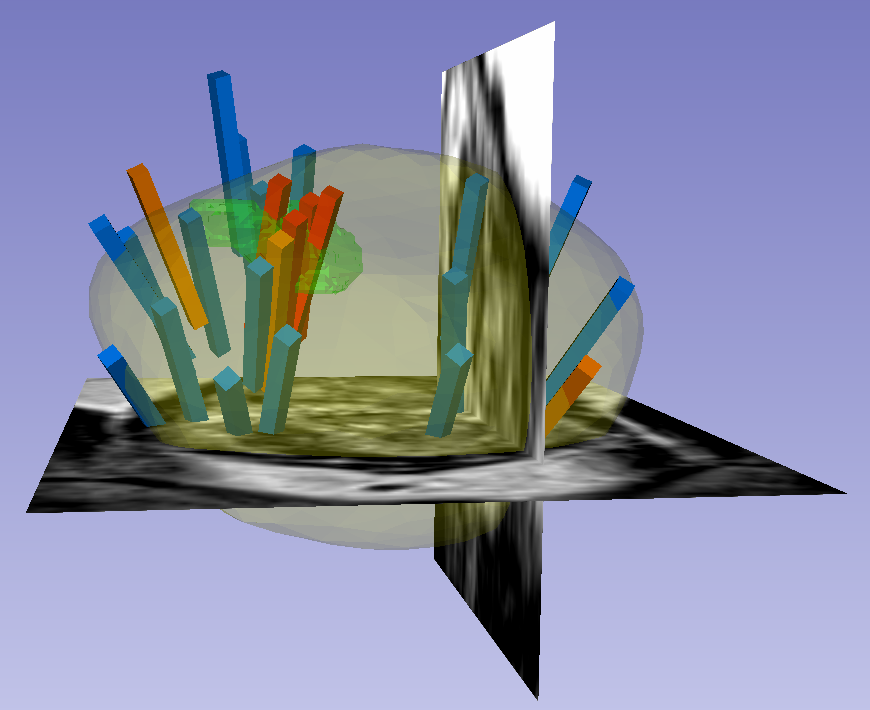
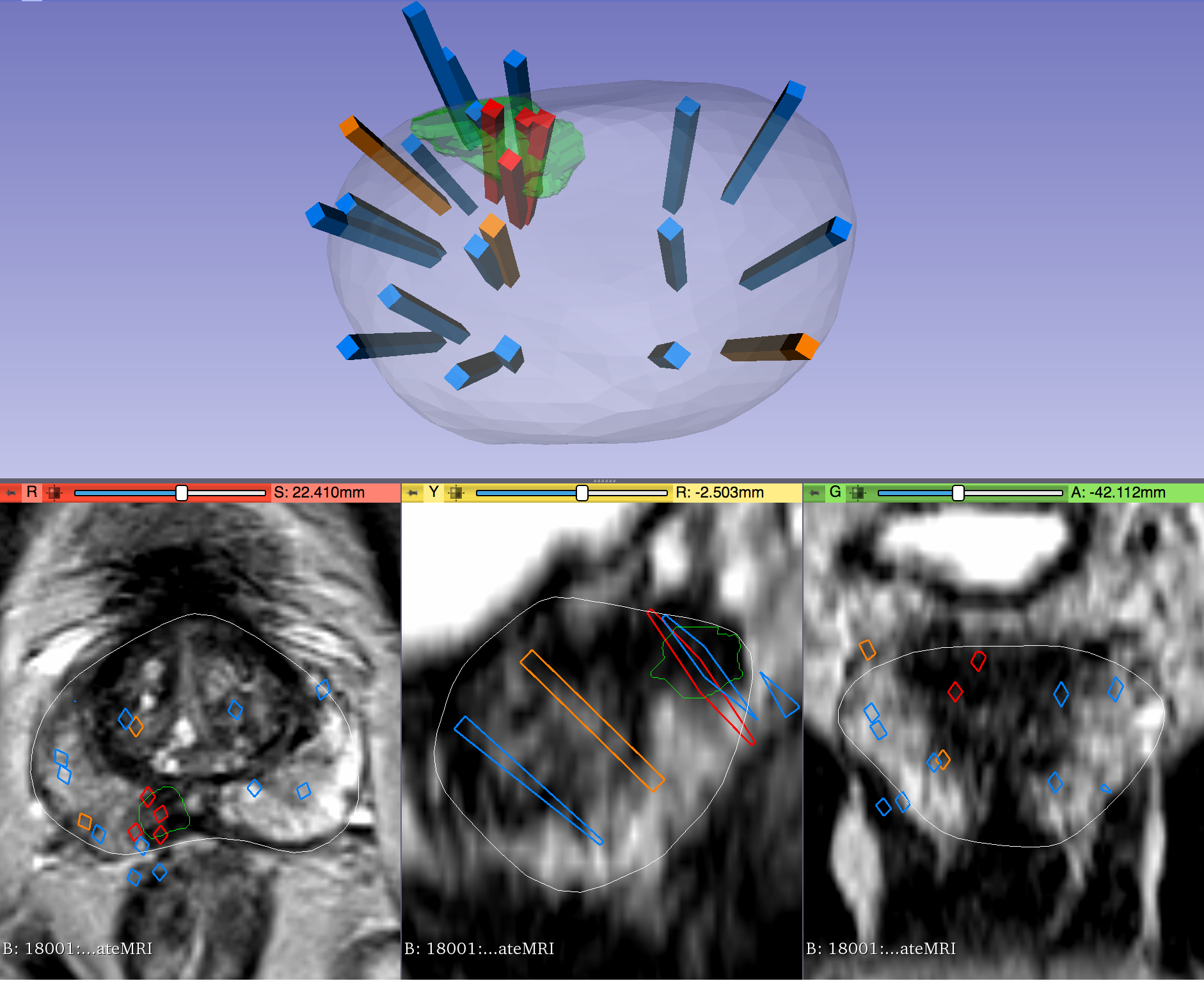
The STL files, which are written in a standard format, can also be imported and visualized with most computer-aided design (CAD) and 3D printing software. Note that, if quantitative analysis is desired, the following transform was applied to US surfaces for 3D slicer compatibility and may need to be reversed for visualization alongside MRI or US images using other software. Transform (which has been applied to all ultrasound data): [0 1 0 0 0 1 1 0 0] Inverse Transform (use to revert ultrasound data to original coordinates): [0 0 1 1 0 0 0 1 0] Instructions for Proper Use of the "Biopsy Overlay" FilesBiopsy vectors, prostate surfaces, and regions of interest can be imported to 3D Slicer in a single step using the collection's "Biopsy Overlay" files. Overlay files associated with ultrasound images are organized into folders with the naming convention "Prostate-MRI-US-Biopsy-XXXX-BXus-seriesUID-####" where "XXXX" is the anonymized patient number and "####" is the series instance UID of corresponding DICOM images. Files associated with MR images have an analogous naming convention "Prostate-MRI-US-Biopsy-XXXX-BXmr-seriesUID-####", but are appended with "(US-date-YYYYMMDD)" where 'YYYYMMDD' is the de-identified acquisition date of the ultrasound series registered to MRI during biopsy. This is specified because in some cases multiple ultrasound-guided biopsies were registered to the same MRI series. Within each folder of "Biopsy Overlays" is a file named "BiopsyVectors.mrml". This can be imported directly into 3D Slicer by either by drag-and-dropping or with File->Add Data. The prostate surface, any ROIs present, and all biopsy vectors will then be displayed within 3D Slicer. The biopsy cores, represented as "line" fiducials, are color-coded as BLUE = benign, ORANGE = Gleason Grade 3+3, and RED = Gleason Grade ≥ 3+4. The prostate is shown as WHITE, and ROIs are GREEN. To visualize these overlays in the context of corresponding MRI or US images, load the corresponding image series to 3D Slicer using File->DICOM. Instructions for Proper Use of the Biopsy and Target SpreadsheetsSpreadsheets accompany the dataset that summarize biopsy and MRI target information. In the spreadsheet "TCIA Biopsy Data," each row represents a single core acquired during ultrasound-guided biopsy. This information includes: - Coordinates of the biopsy core relative to ultrasound images
- Coordinates of the biopsy core relative to MR images. These are present for approximately 70% of the dataset; values of "-1000" are placeholders representing missing data.
- Primary and secondary Gleason score of the biopsy core. Blank entries represent cores containing only benign tissue.
- Length of cancer (mm) within the biopsy core. Blank entries represent cores containing only benign tissue.
- Total tissue length (mm) of the biopsy core; multiple lengths are listed for cores that were fragmented. Tissue lengths are present for approximately 60% of the dataset; blank entries represent cores for which total tissue length was not recorded.
- Core labels (textual description).
- Prostate serum antigen of patient at the time of biopsy.
- Prostate volume of the patient at the time of biopsy, as measured via MRI prostate segmentation.
- De-identified patient number, series instance UID of ultrasound,
| | Localtab |
|---|
| title | Detailed Description |
|---|
| Detailed Description | Image Statistics |
|---|
Modalities | MR, US | Number of Participants | 1151 | Number of Studies | 2799 | Number of Series | 2799 | Number of Images | 61,119 | | Images Size (GB) | 77.6 | Instructions for Proper Use of the STL FilesThe 3D surfaces of this dataset are in stereolithography format (binary representation). They are segmentations representing anatomic regions within the corresponding ultrasound (US) or MRI images. Each file is named "Prostate-MRI-US-Biopsy-XXXX-SURFACETYPE-seriesUID-YYYY.STL" where "XXXX" is the anonymized patient number and "YYYY" is the series instance UID of corresponding DICOM images. "SURFACETYPE" can be one of two identifiers, summarized below. - If "SURFACETYPE" is "ProstateSurface," then the STL file is a segmentation of the prostate gland. These were contoured semi-automatically with Profuse (for MRI) or the Artemis fusion biopsy system (for US), both products of Eigen (Grass Valley, CA). Using these tools, the user manually traced the prostate on one or more image planes, and the remainder of gland was automatically segmented in 3D. The user was then able to adjust vertices of the 3D segmentation as necessary. These segmentations are generally of good quality, though their accuracy is user-dependent.
- If "SURFACETYPE" is "Target," then the STL file is a segmentation of a Region of Interest (ROI) suspicious for harboring prostate cancer. Zero, one, or more than one ROI may be present, and thus are designated "Target1," "Target2," etc. The ROIs corresponding to MRI were traced manually by a radiologist based on their interpretation of the patient's multiparametric MRI study. The ROIs corresponding to US images were acquired by registering the MRI prostate surface nonrigidly and semi-automatically to the US prostate surface. Therefore, the US ROIs are subject to misregistration, with average accuracy of 3-4 mm.
- Note that proximity to an ROI increases the likelihood of, but does not guarantee, prostate cancer. Also note that radiologists assigned each ROI a Likert-like suspicion score of 1-5, with 1 corresponding to very low suspicion of prostate cancer and 5 corresponding to very high suspicion of prostate cancer. These scores, which are roughly equivalent to the "PIRADS Version 2" scoring system, can be found in a spreadsheet included with the dataset.
The STL files can be visualized in 3D Slicer as follows: - Import the surface(s) either by drag-and-dropping or with File->Add Data
- Within the "Models" module, adjust Display->Opacity of the prostate surface in order to visualize any ROI(s) within
- To visualize surfaces in the context of corresponding MRI or US images, load the corresponding image series and then toggle the Slice Display->Visible radio button within the "models" module
- To visualize surfaces in the context of biopsy cores, import the corresponding 3D slicer file named "BiopsyVectors.mrml" by drag-and-dropping or with File->Add Data.
The STL files, which are written in a standard format, can also be imported and visualized with most computer-aided design (CAD) and 3D printing software. Note that, if quantitative analysis is desired, the following transform was applied to US surfaces for 3D slicer compatibility and may need to be reversed for visualization alongside MRI or US images using other software. Transform: [0 1 0 0 0 1 1 0 0] Instructions for Proper Use of the "Biopsy Overlay" FilesBiopsy vectors, prostate surfaces, and regions of interest can be imported to 3D Slicer in a single step using the collection's "Biopsy Overlay" files. Overlay files associated with ultrasound images are organized into folders with the naming convention "Prostate-MRI-US-Biopsy-XXXX-BXus-seriesUID-####" where "XXXX" is the anonymized patient number and "####" is the series instance UID of corresponding DICOM images. Files associated with MR images have an analogous naming convention "Prostate-MRI-US-Biopsy-XXXX-BXmr-seriesUID-####", but are appended with "(US-date-YYYYMMDD)" where 'YYYYMMDD' is the de-identified acquisition date of the ultrasound series registered to MRI during biopsy. This is specified because in some cases multiple ultrasound-guided biopsies were registered to the same MRI series. Within each folder of "Biopsy Overlays" is a file named "BiopsyVectors.mrml". This can be imported directly into 3D Slicer by either by drag-and-dropping or with File->Add Data. The prostate surface, any ROIs present, and all biopsy vectors will then be displayed within 3D Slicer. The biopsy cores, represented as "line" fiducials, are color-coded as BLUE = benign, ORANGE = Gleason Grade 3+3, and RED = Gleason Grade ≥ 3+4. The prostate is shown as WHITE, and ROIs are GREEN. To visualize these overlays in the context of corresponding MRI or US images, load the corresponding image series to 3D Slicer using File->DICOM. Instructions for Proper Use of the Biopsy and Target SpreadsheetsSpreadsheets accompany the dataset that summarize biopsy and MRI target information. In the spreadsheet "TCIA Biopsy Data," each row represents a single core acquired during ultrasound-guided biopsy. This information includes: - Coordinates of the biopsy core relative to ultrasound images
- Coordinates of the biopsy core relative to MR images. These are present for approximately 70% of the dataset; values of "-1000" are placeholders representing missing data.
- Primary and secondary Gleason score of the biopsy core. Blank entries represent cores containing only benign tissue.
- Length of cancer (mm) within the biopsy core. Blank entries represent cores containing only benign tissue.
- Total tissue length (mm) of the biopsy core; multiple lengths are listed for cores that were fragmented. Tissue lengths are present for approximately 60% of the dataset; blank entries represent cores for which total tissue length was not recorded.
- Core labels (textual description).
- Prostate serum antigen of patient at the time of biopsy.
- Prostate volume of the patient at the time of biopsy, as measured via MRI prostate segmentation.
- De-identified patient number, series instance UID of ultrasound, and series instance UID of MR images associated with the biopsy core. Note that MRI UIDs are present for approximately 70% of the dataset; blank entries represent cases for which data was irretrievable or MRI registration was not performed.
In the spreadsheet "Target Data," each row represents a single region of interest (ROI), corresponding to one of the exported Target STL files. This information includes: - Likert-Like "UCLA" score of the ROI. This score predates but is similar to "PIRADSv2." Score values range from 1-5, where 1 is very low suspicion of prostate cancer and 5 is very high suspicion of prostate cancer. Note that scores of 0 or 1 may represent segmentation of non-malignant morphology, or else cases for which a score could not be properly assigned. Only scores of 2 or higher should be considered to correlate with risk of prostate cancer.
- Volume (in CCs) of the ROI.
- Target number of the ROI, enabling correlation between STL surfaces and the suspicion scores for cases with multiple targets.
- De-identified patient number and series instance UID of MR images associated with the ROI.
| Localtab |
|---|
| title | Citations & Data Usage Policy |
|---|
| Citations & Data Usage Policy| Tcia license 4 international |
|---|
| Info |
|---|
| Natarajan, S., Priester, A., Margolis, D., Huang, J., & Marks, L. (2020). Prostate MRI and Ultrasound With Pathology and Coordinates of Tracked Biopsy (Prostate-MRI-US-Biopsy) [Data set]. The Cancer Imaging Archive. DOI: 10.7937/TCIA.2020.A61IOC1A |
| Info |
|---|
| title | Publication Citation |
|---|
| Sonn GA, Natarajan S, Margolis DJ, MacAiran M, Lieu P, Huang J, Dorey FJ, Marks LS. Targeted biopsy in the detection of prostate cancer using an office based magnetic resonance ultrasound fusion device. Journal of Urology 189, no. 1 (2013): 86-91. DOI: 10.1016/j.juro.2012.08.095
|
| Info |
|---|
| Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: 10.1007/s10278-013-9622-7 |
Other Publications Using This DataTCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk. - biopsy core. Note that MRI UIDs are present for approximately 70% of the dataset; blank entries represent cases for which data was irretrievable or MRI registration was not performed.
In the spreadsheet "Target Data," each row represents a single region of interest (ROI), corresponding to one of the exported Target STL files. This information includes: - Likert-Like "UCLA" score of the ROI. This score predates but is similar to "PIRADSv2." Score values range from 1-5, where 1 is very low suspicion of prostate cancer and 5 is very high suspicion of prostate cancer. Note that scores of 0 or 1 may represent segmentation of non-malignant morphology, or else cases for which a score could not be properly assigned. Only scores of 2 or higher should be considered to correlate with risk of prostate cancer.
- Volume (in CCs) of the ROI.
- Target number of the ROI, enabling correlation between STL surfaces and the suspicion scores for cases with multiple targets.
- De-identified patient number and series instance UID of MR images associated with the ROI.
Diffusion-MRI-Supplement-To-Prostate-MRI-US-BiopsyThe following diffusion-weighted MRI series are provided for 837 participants present in the "Prostate-MRI-US-Biopsy" dataset: - Apparent Diffusion Coefficient (ADC)
- High B-value DWI, most commonly with a calculated B-value of 1400
Most commonly these series were generated from DWI images with B-values of [0, 100, 400, 800], consist of 20 image slices, have a resolution of approximately 1.6 x 1.6 x 3.6 mm, and include a window size of the pelvis measuring approximately 256 x 210 mm. Some of the ADC imaging here were created at the scanner, though some were processed later through MATLAB offline to produce a uniform set of ADC pixel data to supplement this collection. Note: Some (0019,xxxx) and (0043,xxxx) tag content may have been mangled in this external processing step, but the images should be consistent. Thus, the T2-weighted MRI series from version 1 of this Collection are supplemented by ADC and high B-value images acquired during the same study, enabling bi-parametric analysis and PI-RADS assessment of regions of interest (ROIs). Scientific questions that could be answered using this data include: - What is the optimum ADC value associated with cancer positivity of prostate biopsy cores
- What is the optimum weighting of diffusion MRI versus T2 in ROI grading (using biopsy as ground-truth for prostate cancer)?
- Can diffusion MRI be used to help an AI model automatically identify and/or segment ROIs suspicious for prostate cancer?
No other public dataset provides both bi-parametric prostate MRI and tracked biopsy pathology. In addition, this data could be of substantial value to participants in the "PI-CAI" challenge, the goal of which is to develop AI models for patient-level diagnosis and lesion-level detection of prostate cancer using bi-parametric MRI. |
| Localtab |
|---|
| title | Citations & Data Usage Policy |
|---|
| Citations & Data Usage Policy| Tcia limited license policy |
|---|
| Info |
|---|
| Natarajan, S., Priester, A., Margolis, D., Huang, J., & Marks, L. (2020). Prostate MRI and Ultrasound With Pathology and Coordinates of Tracked Biopsy (Prostate-MRI-US-Biopsy) (version 2) [Data set]. The Cancer Imaging Archive. DOI: 10.7937/TCIA.2020.A61IOC1A |
| Info |
|---|
| title | Publication Citation |
|---|
| Sonn GA, Natarajan S, Margolis DJ, MacAiran M, Lieu P, Huang J, Dorey FJ, Marks LS. Targeted biopsy in the detection of prostate cancer using an office based magnetic resonance ultrasound fusion device. Journal of Urology 189, no. 1 (2013): 86-91. DOI: 10.1016/j.juro.2012.08.095
PMC3561472
|
| Info |
|---|
| Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: 10.1007/s10278-013-9622-7 |
Additional Publication Resources:The Collection authors suggest the below will give context to this dataset: - Elkhoury, F. F., Felker, E. R., Kwan, L., Sisk, A. E., Delfin, M., Natarajan, S., & Marks, L. S. (2019). Comparison of Targeted vs Systematic Prostate Biopsy in Men Who Are Biopsy Naive. In JAMA Surgery (Vol. 154, Issue 9, p. 811). American Medical Association (AMA). https://doi.org/10.1001/jamasurg.2019.1734
- Jayadevan, R., Felker, E. R., Kwan, L., Barsa, D. E., Zhang, H., Sisk, A. E., Delfin, M., & Marks, L. S. (2019). Magnetic Resonance Imaging–Guided Confirmatory Biopsy for Initiating Active Surveillance of Prostate Cancer. In JAMA Network Open (Vol. 2, Issue 9, p. e1911019). American Medical Association (AMA). https://doi.org/10.1001/jamanetworkopen.2019.11019
- Simopoulos, D. N., Sisk, A. E., Jr., Priester, A., Felker, E. R., Kwan, L., Delfin, M. K., Reiter, R. E., & Marks, L. S. (2019). Cancer core length from targeted biopsy: an index of prostate cancer volume and pathological stage. In BJU International (Vol. 124, Issue 2, pp. 275–281). Wiley. https://doi.org/10.1111/bju.14691
- Kinnaird, A., Sharma, V., Chuang, R., Priester, A., Tran, E., Barsa, D. E., Delfin, M., Kwan, L., Sisk, A., Felker, E., & Marks, L. S. (2020). Risk of Prostate Cancer after a Negative Magnetic Resonance Imaging Guided Biopsy. In Journal of Urology (Vol. 204, Issue 6, pp. 1180–1186). Ovid Technologies (Wolters Kluwer Health). https://doi.org/10.1097/ju.0000000000001232
- Zhou, S. R., Chang, E., Patankar, A., Huang, J., Marks, L. S., & Natarajan, S. (2020). Prostate Cancer Detection Rate of Freehand versus 3-Dimensional Template Mapping Biopsy Using a Magnetic Resonance Imaging-Ultrasound Fusion Device in Biopsy Naïve Men. In Journal of Urology (Vol. 203, Issue 4, pp. 699–705). Ovid Technologies (Wolters Kluwer Health). https://doi.org/10.1097/ju.0000000000000587
- Raman, A. G., Sarma, K. V., Raman, S. S., Priester, A. M., Mirak, S. A., Riskin-Jones, H. H., Dhinagar, N., Speier, W., Felker, E., Sisk, A. E., Lu, D., Kinnaird, A., Reiter, R. E., Marks, L. S., & Arnold, C. W. (2021). Optimizing Spatial Biopsy Sampling for the Detection of Prostate Cancer. In Journal of Urology (Vol. 206, Issue 3, pp. 595–603). Ovid Technologies (Wolters Kluwer Health). https://doi.org/10.1097/ju.0000000000001832
- Brisbane, W. G., Priester, A. M., Ballon, J., Kwan, L., Delfin, M. K., Felker, E. R., Sisk, A. E., Hu, J. C., & Marks, L. S. (2022). Targeted Prostate Biopsy: Umbra, Penumbra, and Value of Perilesional Sampling. In European Urology (Vol. 82, Issue 3, pp. 303–310). Elsevier BV. https://doi.org/10.1016/j.eururo.2022.01.008
- Hassan, Md. R., Islam, Md. F., Uddin, Md. Z., Ghoshal, G., Hassan, M. M., Huda, S., & Fortino, G. (2022). Prostate cancer classification from ultrasound and MRI images using deep learning based Explainable Artificial Intelligence. In Future Generation Computer Systems (Vol. 127, pp. 462–472). Elsevier BV. https://doi.org/10.1016/j.future.2021.09.030
- Padasdao, B., Batsaikhan, Z., Lafreniere, S., Rabiei, M., & Konh, B. (2022). Modeling and Operator Control of a Robotic Tool for Bidirectional Manipulation in Targeted Prostate Biopsy. In 2022 International Symposium on Medical Robotics (ISMR). 2022 International Symposium on Medical Robotics (ISMR). IEEE. https://doi.org/10.1109/ismr48347.2022.9807514
- Priester, A., Fan, R. E., Shubert, J., Rusu, M., Vesal, S., Shao, W., Khandwala, Y. S., Marks, L. S., Natarajan, S., & Sonn, G. A. (2023). Prediction and Mapping of Intraprostatic Tumor Extent with Artificial Intelligence. In European Urology Open Science (Vol. 54, pp. 20–27). Elsevier BV. https://doi.org/10.1016/j.euros.2023.05.018
- Vesal, S., Gayo, I., Bhattacharya, I., Natarajan, S., Marks, L. S., Barratt, D. C., Fan, R. E., Hu, Y., Sonn, G. A., & Rusu, M. (2022). Domain generalization for prostate segmentation in transrectal ultrasound images: A multi-center study. In Medical Image Analysis (Vol. 82, p. 102620). Elsevier BV. https://doi.org/10.1016/j.media.2022.102620
Other Publications Using This DataTCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk. - Hassan, M. R., Islam, M. F., Uddin, M. Z., Ghoshal, G., Hassan, M. M., Huda, S., & Fortino, G. (2022). Prostate cancer classification from ultrasound and MRI images using deep learning based Explainable Artificial Intelligence. Future Generation Computer Systems, 127, 462-472. doi: https://doi.org/10.1016/j.future.2021.09.030
- Kosareva, A. A., Paulenka, D. A., Snezhko, E. V., Bratchenko, I. A., & Kovalev, V. A. (2022). Examining the Validity of Input Lung CT Images Submitted to the AI-Based Computerized Diagnosis. Journal of Biomedical Photonics & Engineering, 8(3). doi:https://doi.org/10.18287/JBPE22.08.030307
- Li, Y., Wang, J., Hu, M., Patel, P., Mao, H., Liu, T., . . . Chen, W. (2023). Prostate gleason score prediction via MRI using capsule network. Paper presented at the Medical Imaging 2023: Computer-Aided Diagnosis.
- Palladino, L., Maris, B., Antonelli, A., & Fiorini, P. (2022). PROST-Net: A Deep Learning Approach to Support Real-Time Fusion in Prostate Biopsy. IEEE Transactions on Medical Robotics and Bionics, 4(2), 323-326. doi: https://doi.org/10.1109/TMRB.2022.3145667
|
| Localtab |
|---|
| Version 2 (Current): 2023/10/20/13| Data Type | Download all or Query/Filter | License |
|---|
| Images (DICOM) 79.4 (GB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/Prostate-MRI-US-Biopsy-NBIA-manifest_v2_20231020.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=Prostate-MRI-US-Biopsy |
|---|
|
|
(Download requires the NBIA Data Retriever) | | Target Data (XLSX, 131 KB) (note corrected to remove 4 mistake rows per PI discussion) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/Target-Data_2019-12-05%20%282%29.xlsx?api=v2 |
|---|
|
|
| | | Biopsy Data (XLSX, 4.25 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/TCIA%20Biopsy%20Data_2020-07-14.xlsx?api=v2 |
|---|
|
|
| | |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYzNSIsInBhc3Njb2RlIjoiNzZkZDBhOWY2Y2YzZjQ1OGQ0ODZlOWE4OTRiOGExMjRjNmNkOGUwNyIsInBhY2thZ2VfaWQiOiI2MzUiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
| stl v1 |
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | | Biopsy Overlays (ZIP, 333 MB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYzMyIsInBhc3Njb2RlIjoiZjY3MzUxMGUyMGY5OGY3YzFhMDE4MDBmZWFjYTRiYWZiYjFjNDE4NCIsInBhY2thZ2VfaWQiOiI2MzMiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
| overlay v1 |
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | |
Added Diffusion-weighted imaging for 837 participants. Version 1: Updated 2020/09/17
| Data Type | Download all or Query/Filter |
|---|
| Images (DICOM, 77.6 GB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/Prostate-MRI-US-Biopsy-NBIA-manifest_20200910.tcia?version=1&modificationDate=1599764668817&api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=Prostate-MRI-US-Biopsy |
|---|
|
|
(Download requires the NBIA Data Retriever) | | Target Data (XLSX, 131 KB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/Target%20Data_2019-12-05.xlsx?api=v2 |
|---|
|
|
| | Biopsy Data (XLSX, 4.25 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/68550661/TCIA%20Biopsy%20Data_2020-07-14.xlsx?api=v2 |
|---|
|
|
| |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYzNSIsInBhc3Njb2RlIjoiNzZkZDBhOWY2Y2YzZjQ1OGQ0ODZlOWE4OTRiOGExMjRjNmNkOGUwNyIsInBhY2thZ2VfaWQiOiI2MzUiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
| stl v1 |
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | Biopsy Overlays (ZIP, 333 MB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYzMyIsInBhc3Njb2RlIjoiZjY3MzUxMGUyMGY5OGY3YzFhMDE4MDBmZWFjYTRiYWZiYjFjNDE4NCIsInBhY2thZ2VfaWQiOiI2MzMiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
| overlay v1 |
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | | Localtab |
|---|
| Version 1 (Current): 2020/09/17| Data Type | Download all or Query/Filter |
|---|
| Images (DICOM) 77.6 (GB) | | | Target Data (XLSX) 131 (KB) | | | Biopsy Data (XLSX) 4.25 (MB) | | | | Biopsy Overlays (ZIP) 333 (MB) |  Image Removed Image Removed
|
|