Detailed Description | |
|---|
Modalities | PT, CT, NM, MR, CR, DX, SC
| Number of Participants | 242 | Number of Studies | 950 | Number of Series | 3,377 | Number of Images | 497,367 | | Image Size (GB) | 145.5 |
Study Accrual: Accrual began in June 2005 and ended in May 2009. Thirty-seven institutions accrued 250 patients to the study. Sixteen patients were ineligible, and eight patients did not have evaluable pretreatment PET, leaving 226 patients. Of these 226 patients, 173 had evaluable post- treatment PET, representing the analysis cohort for the primary end point. Data for 242 of the patients initially enrolled are available in TCIA. Date Offsets: All dates, like the visit date, are protected by presenting just the year; however, dates are also listed as offset days from the base date. The offset dates are used as a means of protecting patient information provided by the local sites in the original data, while allowing users to determine intervals between events. The standard DICOM date tags (i.e. birth dates, imaging study dates, etc.) have been de-identified so that all patients have a baseline study date of January 1, 1960. This falsified date represents the day patients were entered into trial database. The number of days between a subject’s longitudinal imaging studies are accurately preserved. A patient with a study performed on January 4, 1960 means the images were collected 3 days after the base date. For convenience, this calculation has been performed for all scans with the results inserted in DICOM tag (0012,0050) Clinical Trial Time Point ID. This means an imaging study that took place on January 4, 1960 would contain a value of "3" in tag (0012,0050). Imaging Protocol: Conventional modern equipment/techniques for FDG-PET (with or without PET/CT) were used in this study. Patients had to fast for 4 hours and have a blood glucose level less than 200 mg/dL before FDG injection. The FDG dose was not mandated; the recommended dose was 0.14 to 0.21 mCi/kg (approximately 10 to 20 mCi). Emission scanning began 50 to 70 minutes after FDG injection and included the body from upper/mid neck to proximal femurs. Acquisition times for emission and transmission scans were in accordance with the manufacturer’s recommendations. Image Analysis: PET scans were interpreted qualitatively and quantitatively by nuclear medicine physicians/radiologists at each institution, using standardized reporting forms to record the FDG uptake in the primary tumor, regional lymph nodes, and common sites of distant metastasis (ie, bones, adrenals, liver, contralateral lung). These local reviewers were provided with educational materials on image interpretation, specifically describing how to measure peak SUV (SUVpeak). However, formal demonstration of expertise was not mandated. SUVs for regions of interest (ROIs) were determined using two different metrics, maximum SUV (SUVmax) and SUVpeak. SUVmax represented the highest single-voxel SUV within the ROI. SUVpeak represented the mean SUV within a small circular ROI (0.75 to 1.5 cm in diameter) that encompasses the SUVmax voxel. (Thus, SUVpeak will always be lower than SUVmax.). In addition to the institutional interpretations, pre- and post-treatment PET scans were centrally reviewed at ACRIN by an expert nuclear medicine physician with extensive experience in FDG-PET. A single dedicated workstation was used for this purpose, and SUVpeak was measured with an automated program in a circular ROI 1.5 cm in diameter. The central reader was blinded to clinical data and the institutional SUV measurements. Outcomes: Patients were observed for a minimum of 2 years (or until death) after completion of treatment in accordance with standard clinical practice. Non-protocol PET imaging was allowed but not mandated.
Schema  Image Removed Image Removed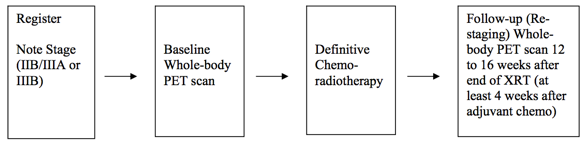 Image Added Image Added
Overview of Clinical Data The basic data flow for legacy ACRIN multi-center clinical trials was that all clinical information provided by the local imaging sites were contained in a series of forms. The form data submitted by local investigators to ACRIN during and after the trial, were manually encoded into the ACRIN CTMS (Clinical Trial Management System), and were cross-checked for accuracy by ECOG-ACRIN personnel. These ACRIN 6668 forms (see the ACRIN 6668 Data Forms page), filled out by the local sites, deliver information on imaging, clinical management of the patient and pathology/outcome variables, like dates of progression and survival, along with other critical information. The image data was initially anonymized while uploading from the local sites through TRIAD software and archived in a DICOM database at ACRIN. After the trial accrual had ended, the clinical data was sent to the Brown statistical center, that is funded by NCI to provide support for ECOG-ACRIN clinical trials, specifically for analysis of the primary and sometimes secondary aims of the trial. The statisticians at Brown strip all the actual dates, names and other PHI from the CTMS data and create a .csv file for each form that has selected information useful for analysis of the trial data. A Form Description file detailing all the forms used in the study accompanies the .csv data files. Additionally, the accompanying Data Dictionary file lists each element for each form that has been selected for data retention along with a description of each form element. Extracting clinical (non-imaging) data example: Beginning with the Form Description.csv file, select the form with the desired information needed, such as form A0.csv the patient Eligibility/Registration form. Next, using the Data Dictionary.csv file, find the form elements listed for A0 (eg., A0exx, where xx is the form element number). The file lists the form number, variable name, its description or label, the type of data, and, when applicable, the option codes and corresponding text values (option code:description pairs like 1=’No’, 2=’Yes’; or 1=’Baseline’, 2=’Post treatment’) for each data element available from the form. In the example in Figure 2, the A0 form element 31d (A0e31d) reports the days between the base date and the day of surgery for the patient. In the corresponding A0.csv file column G lists the days between the base date and surgery for each patient.  Image Removed Image Removed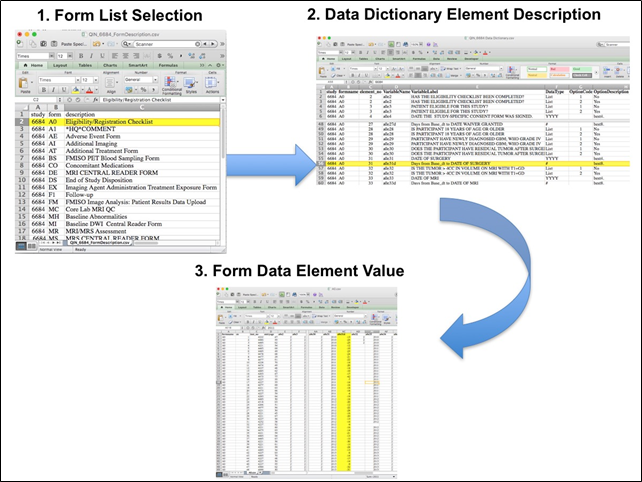 Image Added Image Added In this example of extracting clinical data, the first step is to 1) find the form from the form list, 2) Find the desired element and description in the Data Dictionary and finally 3) extract the values from the .csv data file. The procedure above is basically how the statisticians organized the selected data for export, but the structure of the data dictionaries and individual forms are different for each clinical trial. Much more information about this data set (ACRIN 6668 / RTOG 0235) can be found https://www.acrin.org/6668_protocol.aspx |