Detection, segmentation and classification of nuclei are fundamental analysis operations in digital pathology. Existing state-of-the-art approaches demand extensive amounts of supervised training data from pathologists and may still perform poorly in images from unseen tissue types. We propose an unsupervised approach for histopathology image segmentation that synthesizes heterogeneous sets of training image patches, of every tissue type. Although our synthetic patches are not always of high quality, we harness the motley crew of generated samples through a generally applicable importance sampling method.
This proposed approach, for the first time, re-weighs the training loss over synthetic data so that the ideal (unbiased) generalization loss over the true data distribution is minimized. This enables us to use a random polygon generator to synthesize approximate cellular structures (i.e., nuclear masks) for which no real examples are given in many tissue types, and hence, GAN-based methods are not suited. In addition, we propose a hybrid synthesis pipeline that utilizes textures in real histopathology patches and GAN models, to tackle heterogeneity in tissue textures. Compared with existing state-of-the-art supervised models, our approach generalizes significantly better on cancer types without training data. Even in cancer types with training data, our approach achieves the same performance without supervision cost.
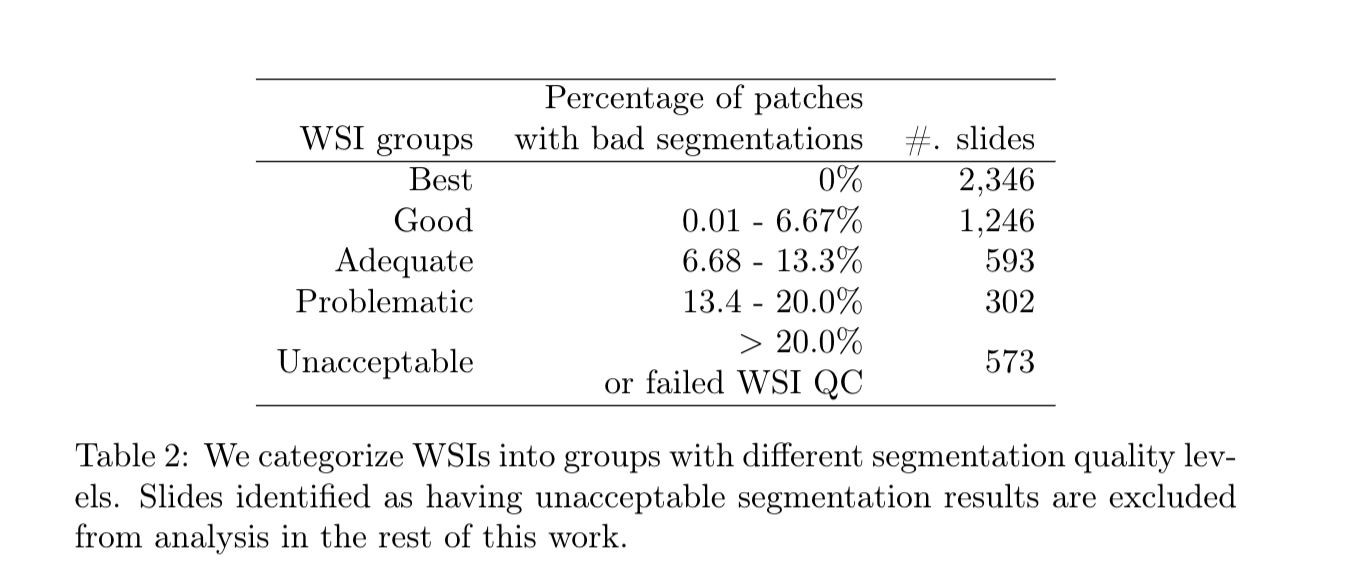
In this dataset we release code and nucleus segmentations in whole slide tissue images with quality control results for over 5000 Whole Slide Images (WSI) in The Cancer Genome Atlas (TCGA) repository. There are two subsets of data: (1) automatic nucleus segmentation data of 5,060 whole slide tissue images of 10 cancer types, with quality control results, and (2) manual nucleus segmentation data of 1,356 image patches from the same 10 cancer types plus additional 4 cancer types.

BLCA Bladder urothelial carcinoma
BRCA Breast invasive carcinoma
CESC Cervical squamous cell carcinoma and endocervical adenocarcinoma
GBM Glioblastoma Multiforme
LUAD Lung adenocarcinoma
LUSC Lung squamous cell carcinoma
PAAD Pancreatic adenocarcinoma
PRAD Prostate adenocarcinoma
SKCM Skin Cutaneous Melanoma
UCEC Uterine Corpus Endometrial Carcinoma
Note that you can also download segmentation data of following 4 cancer types, although they are not officially verified or released.
COAD Colon adenocarcinoma
READ Rectal adenocarcinoma
STAD Stomach adenocarcinoma
UVM Uveal Melanoma
|