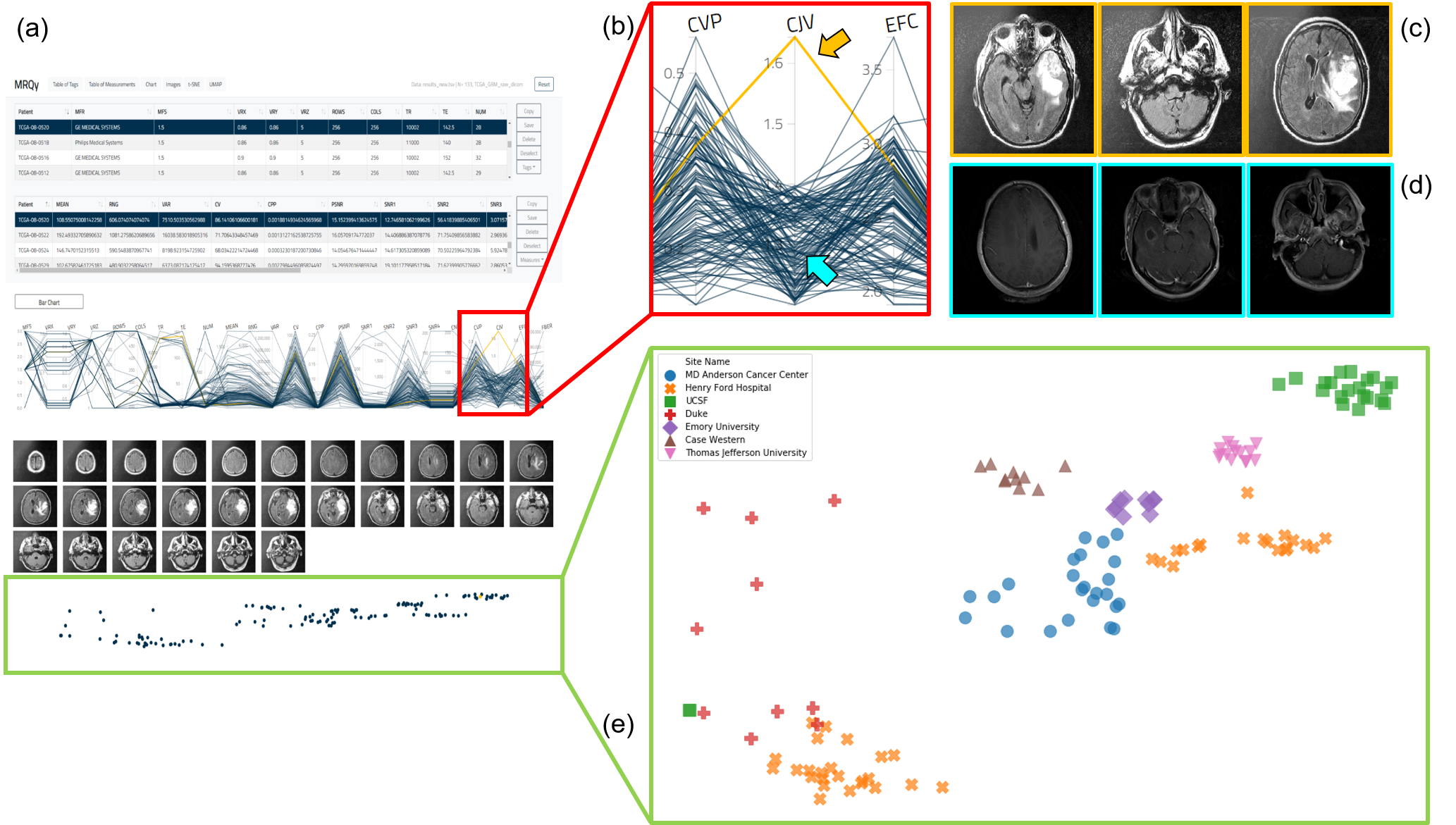
(a) MRQy front-end interface for interrogating TCGA-GBM cohort. (b) Outlier dataset identified on the parallel coordinate chart for the CJV quality measure found to exhibit shading artifacts on (c) representative images, especially when compared to (d) a different dataset without this artifact. (e) t-SNE scatter plot of quality measures revealing presence of site-specific batch effects (colors correspond to different sites, note presence of site-specific clusters).