| Redirect | ||||
|---|---|---|---|---|
|
Summary
| Excerpt |
|---|
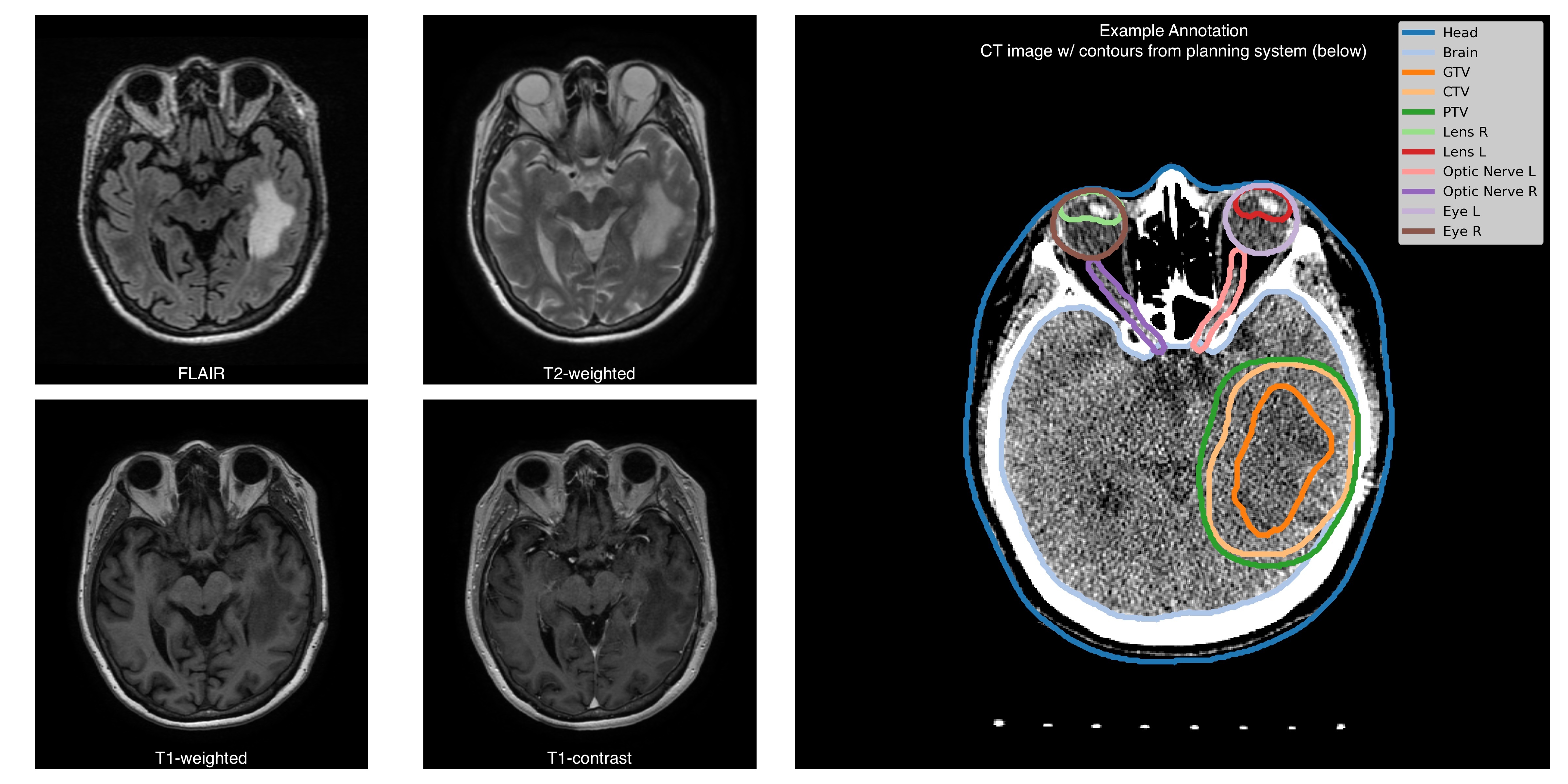
The Burdenko Glioblastoma Progression Dataset (BGPD) is a systematic data collection from 180 patients with primary glioblastoma treated at the Burdenko National Medical Research Center of Neurosurgery between 2014 and 2020. For each patient, the dataset includes imaging studies conducted for radiotherapy planning and follow-up studies. The radiotherapy studies consist of 4 MRI sequences (T1, T1C, T2, FLAIR), a topometric CT scan, and associated radiotherapy planning files (RTSTRUCT, RTPLAN, and RTDOSE). Follow-up studies (from 1 to 8-time per patient) include 2-4 MRI sequences (with a minimal set of T1C and FLAIR) per patient. Additional genetic information (IDH1/2, MGMT mutations); and a treatment response status (tumour progression, tumour pseudoprogression, treatment response) are available for a subset of patients. The MRI studies were obtained from different sites, with scanners from 4 vendors and varying scanning protocols. CT studies were obtained using a unified scanning protocol. Planning and follow-up studiesEach RTSTRUCT contains information on Gross Tumour Volume (GTV), Clinical Target Volume (CTV), and Planning Target Volume (PTV). In addition, each RTSTRUCT includes annotations of 10 anatomical structures: Eyes (Left, Right), Lenses (Left, Right), Optic Nerves (Left, Right), Brain, Brain stem, Chiasm, and external contour (Head). For a subset of patients RTSTRUCTs include annotation of Gross Tumour Volume assessed on follow-up (longitudinal) studies. For each patient, we supplement imaging information with clinical data: IDH1/2 gene mutation is available for 110 patients (97 negative, 13 positive), MGMT promoter methylation status is available for 92 patients (55 negative, 37 positive), death date is available for 80 patients (anonymized with preserved time shift), surgery date (anonymized with preserved time shift) and treatment response status (treatment response, tumour progression, tumour pseudoprogression) are available for all control studies. Additional informationAll MRIs are provided in the original acquisition space, while RTSTRUCTs, RTPLANs, and RTDOSEs are aligned with topometric CT scans. MRI to CT registration files are not provided as a part of the collection, however, here is a github link supporting a containerized solution (written in Python, based on ANTs library) that runs all necessary images and masks alignment. A subset of MRI images are para-transversal (direction cosine vectors as stored in Image Orientation Patient DICOM attribute form an orthonormal basis, but not a canonical one). |
...