...
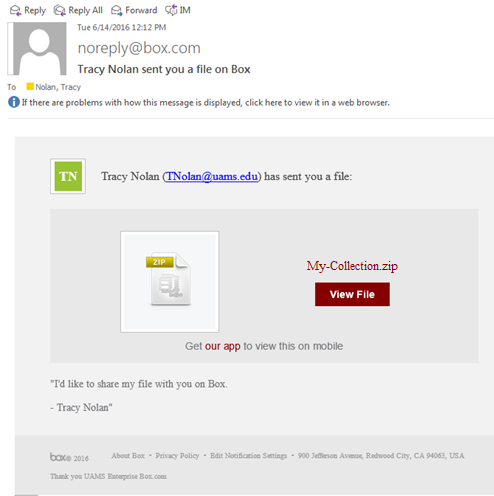
- You will receive an email inviting you to download a zip file. An example of the email follows.
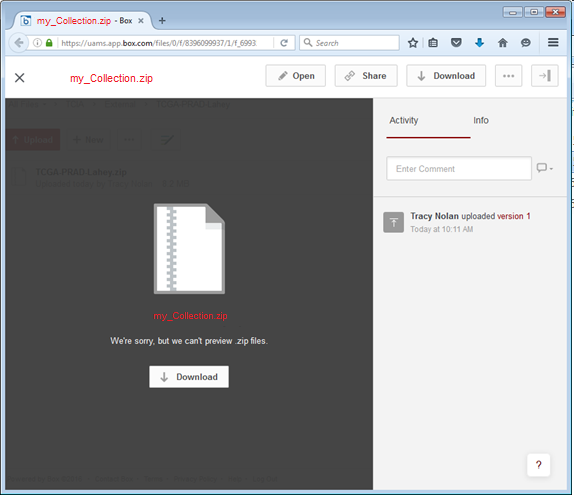
- Click the link to start the download process. The page may read " We're sorry, we can't preview .zip files" that's fine, click the download button as shown:
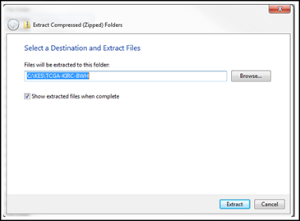
- What you download is compressed. Right click (or apple-click) to choose “Extract” or “Unzip” the downloaded file to a folder where you have write access. You might want to create a folder in the root directory, i.e. ‘C:\CTP’.
Two folders are created. FileSender contains a way to transmit queue DICOM files internally to CTP. CTP contains the anonymization and sendtransmit-to-TCIA transmission software.
Step 2 – Create a mapping table between your patient IDs (probably MRN numbers) and the TCIA IDs
| Note |
|---|
For TCGA cases, it is EXTREMELY important that the mapping is done correctly as these TCGA IDs will be the only link between imaging and tissue samples in the NCI Genomic Data Commons portal. Other Collections with supporting information will also need someone to confirm that dates, File IDs, and PatIDs will be resolved across information types (for example, segmentations, xml, clinical spreadsheets, protein data, histopathology). |
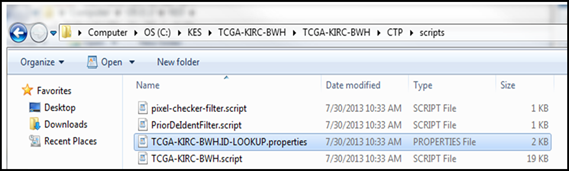
- Navigate to the …/CTP/scripts folder and open the file XXX.ID-LOOKUP.properties file (where XXX represents a name unique to your site) using a plain text editor such as NotePad or WordPad. Please do not use Microsoft Word as that will put in special characters. Example shown below has a lookup table file named
TCGA-KIRC-BWH.ID-LOOKUP.properties. - Complete the Mapping by editing the file you just opened.
- Replace the “Your Patient ID” with the MRN that was assigned to the TCIA ID shown on the right side of the equal sign. An example is shown on the top line. In this fictional case we assigned MRN “01” to TCGA ID TCGA-CZ-9999. If this is not a TCGA collection, then there may be no assigned IDs on the right and you would create your own using the agreed upon naming convention. It is important to use the ptid/ as that is the key to the lookup table.
- For TCGA collections, the pre-populated TCGA IDs represent the possible cases to send. You may not have imaging for all of them.
- Once the mapping is completed, save the file.
...
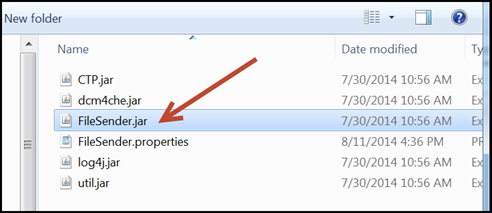
- To use FileSender, open the FileSender folder and double-click
FileSender.jar.
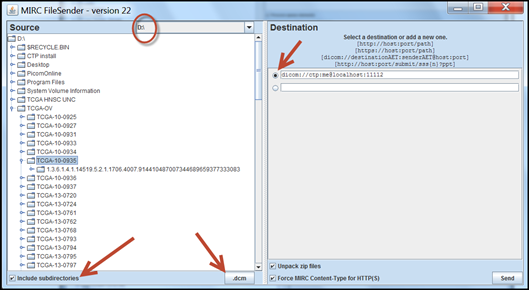
A window appears. - Click the radio button in the destination panel. The line shown is configured to send to CTP on the local computer through port 11112.
A check box at the bottom left can be selected if the DICOM data is in a folder hierarchy - At the lower right of the Source panel is a box that has an *. That just means send all any files. If your images all have extensions of .dcm, you could click the * and change it to .dcm. Only DICOM files will be transmitted, other files will throw an exception.
- Select the directory to the left that has the files you want to send and then click Send.
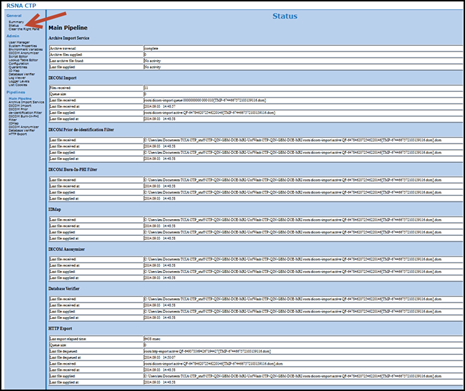
- Monitor the submission process from the Status link on the RSNA CTP homepage.
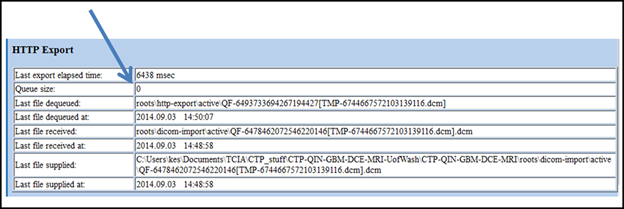
Make sure the last stage “HTTP Export” que queue is empty before stopping CTP.
Info Although the import queue will be empty much sooner, keep CTP running.
...