| Localtab |
|---|
| active | true |
|---|
| title | Data Access |
|---|
| Data Access| Data Type | Download all or Query/Filter |
|---|
Images (DICOM, 1.6 TB) 719 patients |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70230072/ISPY2%20719%20patients%20only%20manifest%20Jan%202022.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=ISPY2 |
|---|
|
|
Note: When DICOM data are downloaded with the NBIA Data Retriever, the app uses "Series Description" values to construct descriptive directory names. In some series for this Collection, characters that are not allowed in directory names are present. So the workaround is to select the “Classic Directory Name” option, which is located above “Select Directory For Downloaded Files.” (Download requires the NBIA Data Retriever) | Images (DICOM, 2.4 TB) I-SPY2 Imaging Cohort 1 dataset, 985 patients |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70230072/ISPY2%20Cohort1%20inclu%20ACRIN6698%20full%20manifest.tcia?api=v2 |
|---|
|
|
| Clinical data (CSV) I-SPY2 Imaging Cohort 1 dataset, 985 patients |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70230072/ISPY2%20Imaging%20Cohort%201%20Clinical%20Data.xlsx?api=v2 |
|---|
|
|
|
Click the Versions tab for more info about data releases. Please contact help@cancerimagingarchive.net with any questions regarding usage. |
| Localtab |
|---|
| title | Detailed Description |
|---|
| Detailed DescriptionImage Statistics |
|
|---|
Modalities | MR,SEG | Number of Patients | 719 | Number of Studies | 2688 | Number of Series | 32411 | Number of Images | 5,586,493 | | Images Size (TB) | 1.6 |
MRI StudiesMRI studies were performed at up to four time points in the course of NAC: - Pre-treatment (T0, optional test/retest visit)
Prior to randomization patients received an MRI study including T2weighted, DWI, and dynamic contrast-enhanced (DCE) acquisitions.
For patients randomized onto a treatment or control arm of I‑SPY 2 subsequent MRI studies with the same required acquisitions were performed: - Early-treatment (T1, optional test/retest visit)
After 3 weeks of regimen 1 treatment (Paclitaxel with or without an experimental agent). - Mid-treatment (T2)
Between Paclitaxel and Anthracycline (AC) treatment regimens - Post-treatment (T3)
Following 4 cycles AC and prior to surgery.
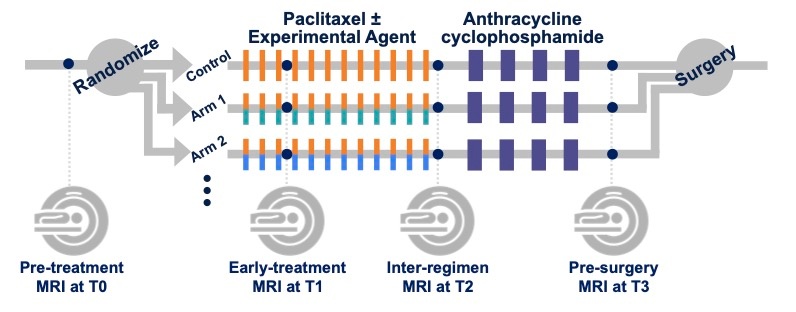
Figure 1. I-SPY 2 study schema. Imaging protocol and included original image seriesMR imaging was performed on a 1.5 or 3.0 Tesla scanner using a dedicated breast radiofrequency coil. All studies for a given patient were required to be performed on the same scanner configuration (model, field strength and breast coil model). Subjects were imaged in the prone position. The image acquisition protocol included a localization scan and two acquisitions: - T2weighted sequence
- T1weighted DCE (80sec < phase duration < 100sec with at least 8 minutes continuous post-injection acquisition)
The DWI acquisitions (b=0, 100, 600, 800 s/mm2 OR b=0, 800) were also acquired but are undergoing quality review and are not included in the collection at this time. Protocol limits on scan parameters for these required acquisitions are given in an accompanying document All required imaging was performed axially with full bilaterally coverage. For the TCIA collection all original images from the two acquisitions are included. Derived images submitted with the study including reformats, CAD software outputs and MIP images are not part of the collection. For the T2weighted acquisitions multiple series are included if they were generated as part of the standard reconstruction process on the scanner. For example, acquisitions utilizing Dixon techniques to create separate fat and water images will have multiple T2weighted series in the collection. Derived object series:Derived objects from the DCE acquisitions are included for the convenience of the user. Some of these are not strictly DICOM compliant and may not be readable by all DICOM software packages. Parametric maps and segmentations are believed to be identical to those utilized in the primary analyses of the I-SPY 2 Trial, but in some cases modifications may have occurred in subsequent processing. DCE “Package” for each study- Single-breast ipsilateral cropped DCE images
- As this was done primarily for computational efficiency, low matrix-sized images (<384 voxels per row) were stored and processed without cropping
- All maps and segmentations are based on these cropped images
- Enhancement maps
- PEearly: Percent enhancement at effective post-contrast time 120-150 sec
- PElate: Percent enhancement at effective post-contrast time ~450 sec
- SER: Signal enhancement ratio SER = PEearly / PElate
Note: SER values were scaled by 1000 to allow integer storage. DICOM software utilizing the rescale slope and intercept fields [ DICOM fields (0028, 1052-1054)] should return the original SER values in the range from 0.0 – 3.0. - All maps are stored as DICOM MRI objects
- Functional Tumor Volume (FTV) analysis mask
- Image storing FTV masking processes on a pixel-by-pixel basis. The mask encodes:
- Pre-contrast background thresholding
- Minimum PE thresholding (70% nom.)
- Manually defined rectangular volume of interest (VOI) for enhancing tumor analysis
- Manually defined “OMIT” regions used to exclude non-tumor enhancing regions that encroached on the rectangular VOI
- Stored as DICOM SEG object in a separate series (see Table 1)
- Download Analysis mask files description for further information:
These DCE derived series can be identified either by Series Description (0008,103e) or by Series Number (0020,0011). See Table 1 for series identification details. Table 1. Series identification for derived DCE objects Data type | Series number | Series Description Match | DCE root directory # | Siemens, GEMS: <root> = 1st original DCE series number Philips: <root> = Original DCE series number / 100 |
|---|
SER Map | <root>1000 | ISPY2: VOLSER: uni-lateral cropped: SER 2 | PE phase <n> Map | <root>100<n> | ISPY2: VOLSER: uni-lateral cropped: PE<n> 2 | Ipsilateral cropped Original | <root>1800 | ISPY2: VOLSER: uni-lateral cropped: original DCE 2 | FTV Analysis Mask1 | <root>1900 | ISPY2: VOLSER: uni-lateral cropped: Analysis Mask 2 |
- FTV Analysis Masks are bit-encoded segmentations encoding all masking steps in the I‑SPY 2 primary FTV analysis. See document Analysis mask files description for a full description of these objects.
- DCE images with less than 384 voxels/row were not cropped for the FTV analysis. For these studies the derived DCE objects are all full bilateral images.
DICOM Dictionary for UCSF Private Attributes in Derived Objects:Calculation parameters and DCE volume analysis results are included in private attributes in some of the derived objects. ACRIN 6698 ISPY2 Shared Private Tag Data Dictionary and ACRIN 6698 ISPY2 DWI and DCE MRI Data Descriptions files are provided for download for researchers wishing to use these parameters and/or results from the primary analyses. Note: the DCE functional tumor volume (FTV) results stored in the SER derived objects are NOT identical to those used in the I‑SPY 2 study due to differences in implementations on different platforms (UCSF in-house software vs. Hologic Inc. AEGIS system). While the differences are generally small and the AEGIS system was validated against the UCSF results in the prior I‑SPY 1 TRIAL, identical results cannot be guaranteed. |
| Localtab |
|---|
| title | Citations & Data Usage Policy |
|---|
| Citations & Data Usage Policy| Tcia license 4 noncommercial |
|---|
| Info |
|---|
| title | Data CitationCitations |
|---|
| - Li, W., Newitt, D. C., Gibbs, J., Wilmes, L. J., Jones, E. F., Arasu, V. A., Strand, F., Onishi, N., Nguyen, A. A.-T., Kornak, J., Joe, B. N., Price, E. R., Ojeda-Fournier, H., Eghtedari, M., Zamora, K. W., Woodard, S. A., Umphrey, H., Bernreuter, W., Nelson, M., … Hylton, N. M. (2022). I-SPY 2 Breast Dynamic Contrast Enhanced MRI (I-SPY2 TRIAL) (Version 1) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.D8Z0-9T85
- Newitt, D. C., Partridge, S. C., Zhang, Z., Gibbs, J., Chenevert, T., Rosen, M., Bolan, P., Marques, H., Romanoff, J., Cimino, L., Joe, B. N., Umphrey, H., Ojeda-Fournier, H., Dogan, B., Oh, K. Y., Abe, H., Drukteinis, J., Esserman, L. J., & Hylton, N. M. (2021). ACRIN 6698/I-SPY2 Breast DWI [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.KK02-6D95
|
| Info |
|---|
| title | Publication Citation |
|---|
| Li, W., Newitt, D. C., Gibbs, J., Wilmes, L. J., Jones, E. F., Arasu, V. A., Strand, F., Onishi, N., Nguyen, A. A.-T., Kornak, J., Joe, B. N., Price, E. R., Ojeda-Fournier, H., Eghtedari, M., Zamora, K. W., Woodard, S. A., Umphrey, H., Bernreuter, W., Nelson, M., … Hylton, N. M. (2020). Predicting breast cancer response to neoadjuvant treatment using multi-feature MRI: results from the I-SPY 2 TRIAL. In npj Breast Cancer (Vol. 6, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/s41523-020-00203-7 |
| Info |
|---|
| Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L., & Prior, F. (2013). The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. In Journal of Digital Imaging (Vol. 26, Issue 6, pp. 1045–1057). Springer Science and Business Media LLC. https://doi.org/10.1007/s10278-013-9622-7 |
Other Publications Using This DataTCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk. |
| Localtab |
|---|
| Version 1 (Current): Updated 2022/05/02| Data Type | Download all or Query/Filter |
|---|
Images (DICOM, 1.6 TB) 719 patients |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70230072/ISPY2%20719%20patients%20only%20manifest%20Jan%202022.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=ISPY2 |
|---|
|
|
Note: When DICOM data are downloaded with the NBIA Data Retriever, the app uses "Series Description" values to construct descriptive directory names. In some series for this Collection, characters that are not allowed in directory names are present. So the workaround is to select the “Classic Directory Name” option, which is located above “Select Directory For Downloaded Files.” (Download requires the NBIA Data Retriever) | Images (DICOM, 2.4 TB) I-SPY2 Imaging Cohort 1 dataset, 985 patients |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70230072/ISPY2%20Cohort1%20inclu%20ACRIN6698%20full%20manifest.tcia?api=v2 |
|---|
|
|
| Clinical data (CSV) I-SPY2 Imaging Cohort 1 dataset, 985 patients |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70230072/ISPY2%20Imaging%20Cohort%201%20Clinical%20Data.xlsx?api=v2 |
|---|
|
|
|
|
|