Description
In short, this publication This data applies a radiomic approach to computed tomography data of 1,019 patients with lung or head-and-neck cancer which are described in Nature Communications (http://doi.org/10.1038/ncomms5006). The various arms of the study are represented in TCIA as distinct Collections including NSCLC-Radiomics (Lung1), NSCLC-Radiomics-Genomics (Lung3), Head-Neck-Radiomics-HN1 (H&N1), NSCLC-Radiomics-Interobserver1 (Multiple delineation), and RIDER Lung CT Segmentation Labels from: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach (RIDER test/retest).
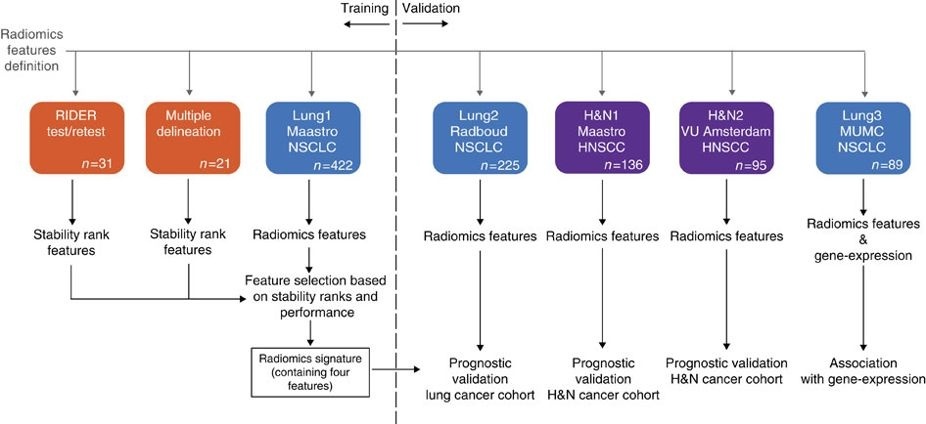 Image Added
Image Added
Radiomics refers to the comprehensive quantification of tumour phenotypes by applying a large number of quantitative image features. In present analysis 440 features quantifying tumour image intensity, shape and texture, were extracted. We found that a large number of radiomic features have prognostic power in independent data sets, many of which were not identified as significant before. Radiogenomics analysis revealed that a prognostic radiomic signature, capturing intra-tumour heterogeneity, was associated with underlying gene-expression patterns. These data suggest that radiomics identifies a general prognostic phenotype existing in both lung and head-and-neck cancer. This may have a clinical impact as imaging is routinely used in clinical practice, providing an unprecedented opportunity to improve decision-support in cancer treatment at low cost.
...
| Localtab Group |
|---|
| Localtab |
|---|
| active | true |
|---|
| title | Data Access |
|---|
| Data AccessNote: This data is restricted for commercial use. Please contact Hugo Aerts, hugo_aerts@dfci.harvard.edu with any questions on usage.Click the Download button to save a ".tcia" manifest file to your computer, which you must open with the NBIA Data Retriever | Data Type | Download all or Query/Filter |
|---|
| Image Data (DICOM) | | | Clinical Data (CSV, XLS) | | | ) and Clinical Data | Please refer to each Collection page to download available images and clinical data: | NSCLC-Radiomics-Genomics (Lung3) Gene Expression Data | | Gene Expression Data | |
Please contact help@cancerimagingarchive.net with any questions regarding usage. |
| Localtab |
|---|
| title | Citations & Data Usage Policy |
|---|
| Citations & Data Usage Policy These collections are freely This collection may not be used for commercial purposes. It is available to browse, download, and use for commercial, scientific and educational purposes as outlined in the Creative Commons Attribution 3.0 Unported Licensethe Attribution-NonCommercial 3.0 Unported (CC BY-NC 3.0) https://creativecommons.org/licenses/by-nc/3.0/. See TCIA's Data Usage Policies and Restrictions for additional details. Questions may be directed to help@cancerimagingarchive.net. Please be sure to acknowledge both this data set and TCIA in publications by including the following citations in your work: | Info |
|---|
| Hugo J. W. L. Aerts; Emmanuel Rios Velazquez; Ralph T. H. Leijenaar; Chintan Parmar; Patrick Grossmann; Sara Cavalho; Johan Bussink; René Monshouwer; Benjamin Haibe-Kains; Derek Rietveld; Frank Hoebers; Michelle M. Rietbergen; C. René Leemans; Andre Dekker; John Quackenbush; Robert J. Gillies; Philippe Lambin. (2014). Data from: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2014..UA0JGPDG |
| Info |
|---|
| Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. (paper) |
In addition to the dataset citation above, please be sure to cite the following if you utilize these data in your research: | Info |
|---|
| title | Publication Citation |
|---|
| Aerts, H. J. W. L., Velazquez, E. R., Leijenaar, R. T. H., Parmar, C., Grossmann, P., Cavalho, S., … Lambin, P. (2014, June 3). Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nature Communications. Nature Publishing Group. http://doi.org/10.1038/ncomms5006 |
Other Publications Using This DataTCIA maintains a list of publications that leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk. |
| Localtab |
|---|
| Version 2 (Current): 2020/03/23Added links to the recently published TCIA collections which reflect the additional arms of the study described in Nature Communications (http://doi.org/10.1038/ncomms5006).
| Data Type | Download all or Query/Filter |
|---|
| Image Data (DICOM) and Clinical Data | Please refer to each Collection page to download available images and clinical data: | NSCLC-Radiomics-Genomics (Lung3) Gene Expression Data | |
Version 1 (Current): 2016/08/02| Data Type | Download all or Query/Filter |
|---|
| Image Data (DICOM) | | | Clinical Data (CSV, XLS) | | | Gene Expression Data | |
|
|
