| Localtab |
|---|
| active | true |
|---|
| title | Data Access |
|---|
| Data Access| Data Type | Download all or Query/Filter | License |
|---|
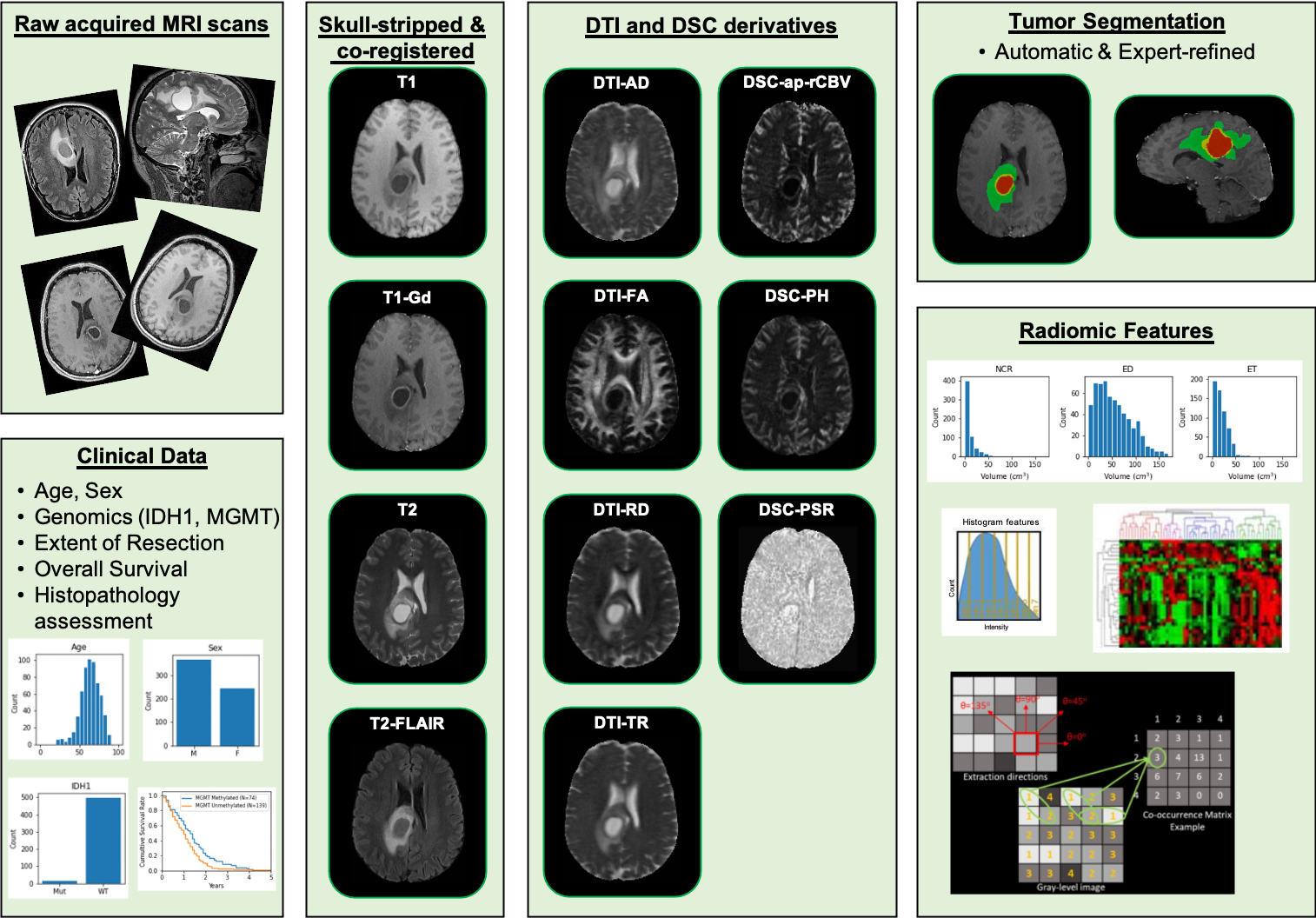
Images |
, Segmentations, and Radiation Therapy Structures/Doses/Plans XXX << latter two items only if DICOM SEG/RTSTRUCT/RTDOSE/PLAN exist >>  Image Removed Image Removed Image Removed Image Removed |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_DownloadManifest20221129.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=UPENN-GBM |
|---|
|
|
(Download requires the NBIA Data Retriever) |
Tissue Slide SVS, XX.X GB) Image Removed Image Removed Image Removed Image Removed
| | Clinical data (CSV) |  Image Removed Image Removed
| Genomics (web) | NIfTI, 69 GB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYwNCIsInBhc3Njb2RlIjoiYzJiMjI2Mzg5ZjljYWE0NWNkYjc4MzM4NWE4Yzc2MjBjNGU1NDY1MiIsInBhY2thZ2VfaWQiOiI2MDQiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | Histopathology Images (NDPI, 149 GB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjUwOSIsInBhc3Njb2RlIjoiNTkyMmNjY2U5Nzc5N2YxZTQ2NGIwMWNiMjY0NzQzMjg3YjY4MDA2NiIsInBhY2thZ2VfaWQiOiI1MDkiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://pathdb.cancerimagingarchive.net/imagesearch?f[0]=collection:upenn_gbm |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | | Clinical Data (CSV, 51 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_clinical_info_v1.2.csv?api=v2 |
|---|
|
|
| | | Histopathology to Radiology Filename Mapping (CSV, 2 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/radiology_mapping.csv?version=2&modificationDate=1666634570946&api=v2 |
|---|
|
|
| | | Image acquisition parameters (CSV, 195 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_acquisition.csv?api=v2 |
|---|
|
|
| | | Data availability per subject (CSV, 126 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_data_availability.csv?api=v2 |
|---|
|
|
| | | CaPTk radiomic features list (CSV, 6 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_CaPTk_radiomic_features_list.csv?api=v2 |
|---|
|
|
| | | CaPTk radiomic feature parameter file (CSV, 4 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_CaPTk_fe_params.csv?api=v2 |
|---|
|
|
| | | Radiomic Data (ZIP,15.37 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/radiomic_features_CaPTk.zip?api=v2 |
|---|
|
|
| |
 Image Removed Image RemovedClick the Versions tab for more info about data releases. Please contact help@cancerimagingarchive.net with any questions regarding usage. | Nci_crdc additional resources |
|---|
|
| Localtab |
|---|
| title | Detailed Description |
|---|
| Detailed DescriptionImage Statistics |
|
|
|---|
Modalities | MR | Pathology | Number of Patients | 630 | 34 | Number of Studies | 3,301 | N/A | Number of Series | 3,680 | N/A | Number of Images | 828,234 | 71 | | Images Size (GB) |
Note from the submitting group: The NIfTI images are all registered to a common atlas (SRI) using a uniform preprocessing and the segmentation are aligned with them. Therefore the NIfTI images will not align with the DICOM images, by design. If you load the NIfTI images (like T1/T2) and their related segmentation, these will line up. |
| Localtab |
|---|
| title | Citations & Data Usage Policy |
|---|
| Citations & Data Usage PolicyAdd any special restrictions in here. These collections are freely available to browse, download, and use for commercial, scientific and educational purposes as outlined in the Creative Commons Attribution 4.0 International License. Questions may be directed to help@cancerimagingarchive.net. Please be sure to acknowledge both this data set and TCIA in publications by including the following citations in your work: | Info |
|---|
| DOI goes here. Create using Datacite with information from Collection Approval form |
| Info |
|---|
| title | Publication Citation |
|---|
| We ask on the proposal form if they have ONE traditional publication they'd like users to cite. |
| Tcia limited license policy |
|---|
| Info |
|---|
| Bakas, S., Sako, C., Akbari, H., Bilello, M., Sotiras, A., Shukla, G., Rudie, J. D., Flores Santamaria, N., Fathi Kazerooni, A., Pati, S., Rathore, S., Mamourian, E., Ha, S. M., Parker, W., Doshi, J., Baid, U., Bergman, M., Binder, Z. A., Verma, R., … Davatzikos, C. (2021). Multi-parametric magnetic resonance imaging (mpMRI) scans for de novo Glioblastoma (GBM) patients from the University of Pennsylvania Health System (UPENN-GBM) (Version 2) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.709X-DN49 |
| Info |
|---|
| title | Publication Citation |
|---|
| Bakas, S., Sako, C., Akbari, H., Bilello, M., Sotiras, A., Shukla, G., Rudie, J. D., Flores Santamaria, N., Fathi Kazerooni, A., Pati, S., Rathore, S., Mamourian, E., Ha, S. M., Parker, W., Doshi, J., Baid, U., Bergman, M., Binder, Z. A., Verma, R., Lustig, R., Desai, A. S., Bagley, S. J., Mourelatos, Z., Morrissette, J., Watt, C. D., Brem, S., Wolf, R. L., Melhem, E. R., Nasrallah, M. P., Mohan, S., O’Rourke, D. M., Davatzikos, C. (2022). The University of Pennsylvania glioblastoma (UPenn-GBM) cohort: advanced MRI, clinical, genomics, & radiomics. In Scientific Data (Vol. 9, Issue 1). https://doi.org/10.1038/s41597-022-01560-7 | | Info |
|---|
| Only if they ask for special acknowledgments like funding sources, grant numbers, etc in their proposal. |
| Info |
|---|
| Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L., & Prior, F. (2013). The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, . Journal of Digital Imaging, Volume 26, Number (6, December, 2013, pp 1045-1057. DOI: ), 1045–1057. https://doi.org/10.1007/s10278-013-9622-7 |
Other Publications Using This DataTCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk. |
| Localtab |
|---|
| Version X 2 (Current): Updated yyyy/mm/dd2022/10/24Note: 2023/12/5: updated clinical data file (v1.2) to include more information about censor/survival. | Data Type | Download all or Query/Filter | License |
|---|
Images (DICOM, |
xxx GB) Image Removed Image Removed Image Removed Image Removed
(Requires NBIA Data Retriever.) | | Clinical Data (CSV) | Link | | Other (format) |  Image Removed Image Removed
| 4 GB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_DownloadManifest20221129.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=UPENN-GBM |
|---|
|
|
(Download requires the NBIA Data Retriever) | | Images (NIfTI, 69 GB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYwNCIsInBhc3Njb2RlIjoiYzJiMjI2Mzg5ZjljYWE0NWNkYjc4MzM4NWE4Yzc2MjBjNGU1NDY1MiIsInBhY2thZ2VfaWQiOiI2MDQiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | Histopathology Images (NDPI, 149 GB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjUwOSIsInBhc3Njb2RlIjoiNTkyMmNjY2U5Nzc5N2YxZTQ2NGIwMWNiMjY0NzQzMjg3YjY4MDA2NiIsInBhY2thZ2VfaWQiOiI1MDkiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://pathdb.cancerimagingarchive.net/imagesearch?f[0]=collection:upenn_gbm |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | | Clinical Data (CSV, 51 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_clinical_info_v1.2.csv?api=v2 |
|---|
|
|
| | | Histopathology to Radiology Filename Mapping (CSV, 2kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/radiology_mapping.csv?version=1&modificationDate=1666627440627&api=v2 |
|---|
|
|
| | | Image acquisition parameters (CSV, 195 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_acquisition.csv?api=v2 |
|---|
|
|
| | | Data availability per subject (CSV, 126 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_data_availability.csv?api=v2 |
|---|
|
|
| | | CaPTk radiomic features list (CSV, 6 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_CaPTk_radiomic_features_list.csv?api=v2 |
|---|
|
|
| | | CaPTk radiomic feature parameter file (CSV, 4 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_CaPTk_fe_params.csv?api=v2 |
|---|
|
|
| | | Radiomic Data (ZIP,15.37 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/radiomic_features_CaPTk.zip?api=v2 |
|---|
|
|
| |
changes: Histopathology NDPI slides added to collection. CSV file for mapping Radiology subject IDs to Histopathology patient and image IDs where available (note: not all Radiology data has associated pathology data and vice versa). Version 1: 2022/06/21| Data Type | Download all or Query/Filter | License |
|---|
Images (DICOM, 139.4 GB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM-manifest_20210923.tcia?api=v2 |
|---|
|
|
| Tcia button generator |
|---|
| label | Search |
|---|
| url | https://nbia.cancerimagingarchive.net/nbia-search/?MinNumberOfStudiesCriteria=1&CollectionCriteria=UPENN-GBM |
|---|
|
|
(Download requires the NBIA Data Retriever) | | Images (NIfTI, 69 GB) |
| Tcia button generator |
|---|
| url | https://faspex.cancerimagingarchive.net/aspera/faspex?context=eyJyZXNvdXJjZSI6InBhY2thZ2VzIiwidHlwZSI6ImV4dGVybmFsX2Rvd25sb2FkX3BhY2thZ2UiLCJpZCI6IjYwNCIsInBhc3Njb2RlIjoiYzJiMjI2Mzg5ZjljYWE0NWNkYjc4MzM4NWE4Yzc2MjBjNGU1NDY1MiIsInBhY2thZ2VfaWQiOiI2MDQiLCJlbWFpbCI6ImhlbHBAY2FuY2VyaW1hZ2luZ2FyY2hpdmUubmV0In0= |
|---|
|
|
(Download and apply the IBM-Aspera-Connect plugin to your browser to retrieve this faspex package) | | | Clinical Data (CSV, 51 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_clinical_info_v1.0.csv?api=v2 |
|---|
|
|
| | | Image acquisition parameters (CSV, 195 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_acquisition.csv?api=v2 |
|---|
|
|
| | | Data availability per subject (CSV, 126 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_data_availability.csv?api=v2 |
|---|
|
|
| | | CaPTk radiomic features list (CSV, 6 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_CaPTk_radiomic_features_list.csv?api=v2 |
|---|
|
|
| | | CaPTk radiomic feature parameter file (CSV, 4 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/UPENN-GBM_CaPTk_fe_params.csv?api=v2 |
|---|
|
|
| | | Radiomic Data (ZIP,15.37 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/radiomic_features_CaPTk.zip?api=v2 |
|---|
|
|
| | | CaPTk radiomic feature parameter (CSV, 5KB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/70225642/captk_parameter_file_ispy1.csv?api=v2 |
|---|
|
|
| |
|
|