PROSTATEx Challenge (November 21, 2016 to February 16, 2017)SPIE, along with the support of the American Association of Physicists in Medicine (AAPM) and the National Cancer Institute (NCI), conducted a “Grand Challenge” on quantitative image analysis methods for the diagnostic classification of clinically significant prostate lesions. For more details, go to https://prostatex.grand-challenge.org/ . Training and test cohorts along with supplemental Ktrans images and lesion information can be obtained separately using the following options for download:
| Data Type | Training Cohort (204 subjects) | Test Cohort (140 subjects) |
|---|
Download images (DICOM, 126 kB) (DICOM, 121 kB)
|
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/PROSTATEx-train.tcia?version=1&modificationDate=1534787030035&api=v2 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/PROSTATEx-test.tcia?version=1&modificationDate=1534787450806&api=v2 |
|---|
|
|
| Download Ktrans images (.mhd 161 kB) (.mhd 119 kB) |
| Tcia button generator |
|---|
| url | https://app.box.com/s/y871i386j4o9rqwcsms5mqni63nojzx1 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://app.box.com/s/k3iofc0r3ktjnb4f7105lssdzjon95ie |
|---|
|
|
| Download lesion information (.zip, 281 kB) (zip, 236 kB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateX-TrainingLesionInformationv2.zip?version=2&modificationDate=1483479231532&api=v2 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateX-TestLesionInformation.zip?version=2&modificationDate=1483479234096&api=v2 |
|---|
|
|
| Download lesion reference thumbnails (.bmp, 37.6 MB) (.bmp, 23.3 MB) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateX-Screenshots-Train.zip?version=1&modificationDate=1479401241653&api=v2 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://app.box.com/s/7jn4jtd3pbi9rlurc4pvnkx554lnfljw |
|---|
|
|
|
The images come in two encodings. The acquired MR is provided in DICOM encoding. Additionally Ktrans images are provided. They come in mhd format. Ktrans is a key pharmacokinetic parameter computed from the available Dynamic contrast enhanced T1-weighted series. Each patient has one study with several DICOM images and one Ktrans image. The Ktrans image is encoded in two files ProstateX-[ProxID]-Ktrans.[mhd/zraw], where ProxID is the ProstateX patient identifier. The DICOM images comprise several Series each comprising several Instances. The DICOM files are documented in the ProstateX-Images.csv file. The columns in that file encode the following: ProxID – ProstateX patient identifier. Name – Series Description Studydate – Study Date fid – Finding ID Pos – Scanner Coordinate position of the finding WorldMatrix – Matrix describing image orientation and scaling ijk – image col,row,slice coordinate of finding ImageUID – Image Identifier TopLevel 0 - Series forms one image 1 – A set of Series forms a 4D image (e.g. Dynamic MR) NA – Series form one image, but is part of a Level 1 4D image
SpacingBetweenSlices – Scalar Spacing between slices VoxelSpacing – Vector with x,y,z spacing scalars Dim – Vector with 4D dimensions of the image DCMSerDescr – The original DICOM Series Description DCMSerUID – The DICOM Series UID DCMSerNum – The DICOM Series Number InstanceUIDList – DICOM Instances that make up this series ImageUIDList – TopLevel-NA Images the make up this Toplevel 1 image
For example, to get the ADC image of Patient ProstateX-0123 do the following. After you imported the DICOM files into your environment, go to patient ProstateX-0123 and find the series with ADC in it. In this case it is ‘ep2d_diff_tra_DYNDIST_ADC’. It has SeriesNumber 8. The DICOM images in that series form the ADC image for this challenge. Image slice j at coordinate i,j contains a finding fid. See findings for more details. Findings The findings are documented in the ProstateX-Findings.csv table. Documentation for the columns in that table is as follows: pos - Scanner Coordinate position of the finding ClinSig – Identifier available in training set that identifies whether this is a clinically significant finding. Either the biopsy GleasonScore was 7 or higher. Findings with a PIRADS score 2 were not biopsied and are not considered clinically significant. In our center the occurrence of clinically significant cancer in PIRADS 2 lesions is less than 5%.
Note, ProxID is the PROSTATEx case ID, and fid is the finding (i.e., lesion) ID in both the ProstateX-Findings.csv file and the ProstateX-Images.csv file. The Findings spreadsheet has one row per lesion (if a case has only one lesion, then the only fid for that case will be “1” , alternately if a case has two lesions, then there will be an fid of “1” and an fid of “2” for that case). The Images spreadsheet has a row for every image that contains the lesion - that is why there are multiple rows with the same (ProxID, fid) combination.
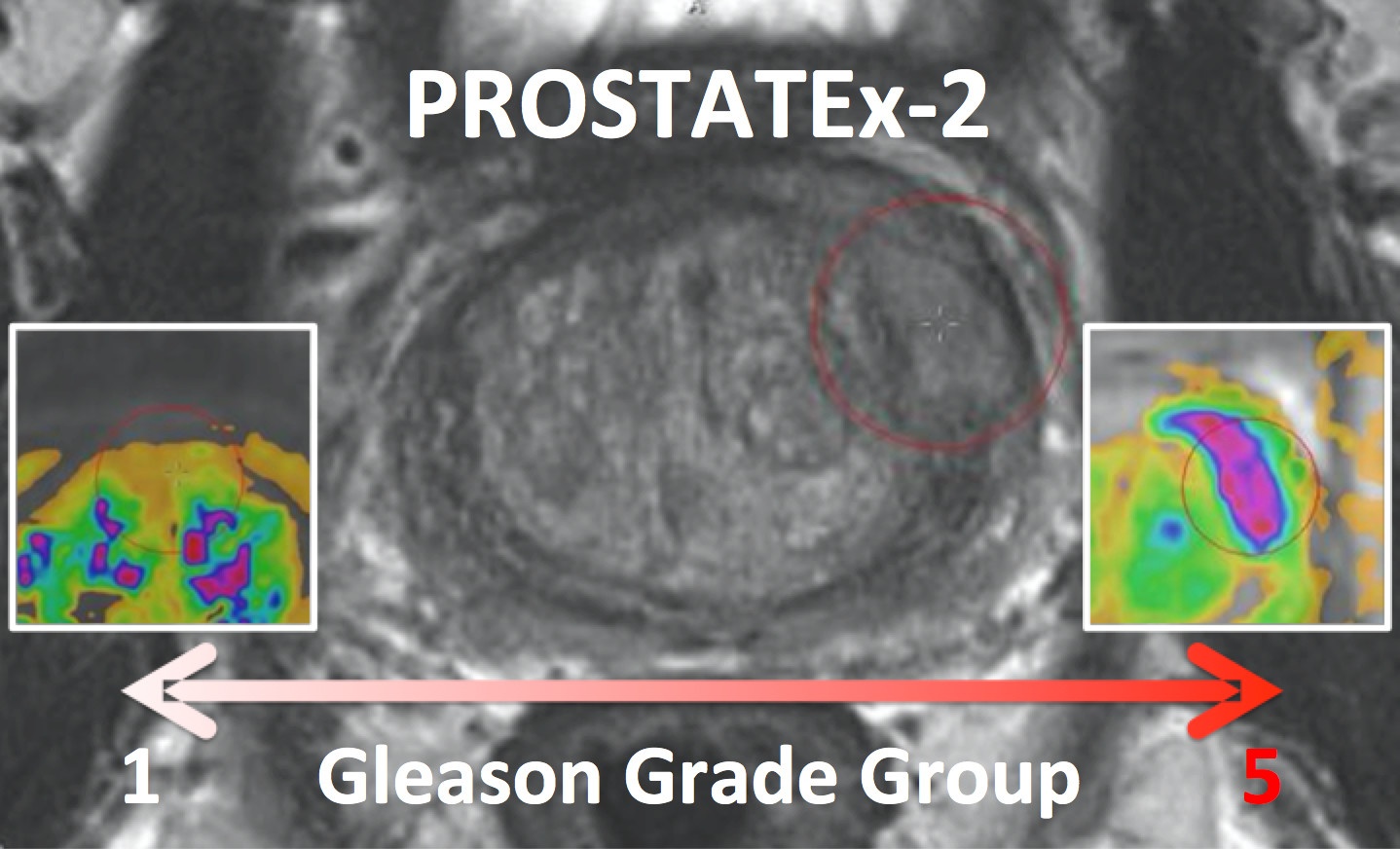
PROSTATEx-2 — SPIE-AAPM-NCI Prostate MR Gleason Grade Group Challenge (May 15, 2017 to August 3, 2017)The American Association of Physicists in Medicine (AAPM), along with the SPIE (the international society for optics and photonics) and the National Cancer Institute (NCI), conducted a part 2 “Grand Challenge” on the development of quantitative multi-parametric magnetic resonance imaging (MRI) biomarkers for the determination of Gleason Grade Group in prostate cancer. For more details about PROSTATEx-2 please go to http://www.aapm.org/GrandChallenge/PROSTATEx-2/default.asp . Training and test cohorts along with supplemental Ktrans images and lesion information can be obtained separately using the following options for download:
| Data Type | Training Cohort (112 subjects) | Test Cohort (70 subjects) |
|---|
Download images (DICOM) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/PROSTATExChallenge2-v2-doiJNLP.tcia?version=1&modificationDate=1534787028529&api=v2 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/PROSTATEx2-test.tcia?version=1&modificationDate=1534787027933&api=v2 |
|---|
|
|
| Download Ktrans images (.mhd) |
| Tcia button generator |
|---|
| url | https://app.box.com/s/xv6j8yhq4nd7c1eqdn88n1maw3un0xbv |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://app.box.com/s/ibue53ch74tst8hd7hknvhw01jxruw2v |
|---|
|
|
| Download lesion information (.zip) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateX2-DataInfo-Train.zip?version=1&modificationDate=1494608672116&api=v2 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateX2-DataInfo-Test.zip?version=1&modificationDate=1496674918589&api=v2 |
|---|
|
|
| Download lesion reference thumbnails (.bmp) |
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateXChallenge2ScreenshotsTrain.zip?version=1&modificationDate=1494518725752&api=v2 |
|---|
|
|
|
| Tcia button generator |
|---|
| url | https://wiki.cancerimagingarchive.net/download/attachments/23691656/ProstateXChallenge2ScreenshotsTest.zip?version=1&modificationDate=1496674921348&api=v2 |
|---|
|
|
|
The training set will consist of 112 findings. This dataset will be representative of the technical properties (scanner type, acquisition parameters, file format) and the nature of the prostate lesions of the test set. An associated Excel file will include case name, the coordinates of the centroid of all lesions, and the lesion ‘truth’ label (Gleason Grade Group). Reference thumbnail images of the lesions will also be provided. The test set will consist of 70 findings. The locations of the lesions will be specified in the accompanying Excel file that will follow the same format as for the training set with the omission of the ‘truth’ labels. Reference thumbnail images of the lesions will also be provided. |