Detailed Description | |
|---|
Modalities | MR | Number of Patients | 346 | Number of Studies | 349 | Number of Series | 18,321 | Number of Images | 309,251 | | Images Size (GB) | 15.1 |
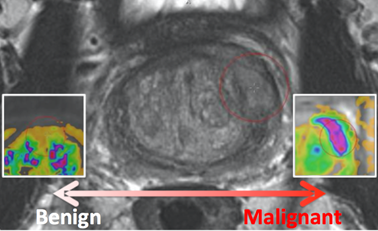
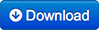
Prostate MR in accordance with ACR PIRADS2.0 comprises at least 3 types of images or parameters that should be jointly analyzed for the assessment of prostate cancer. The prostate MR imaging was performed at the Radboud University Medical Centre (Radboudumc) in the Prostate MR Reference Center under supervision of prof. dr. Barentsz. The Radboudumc is located in Nijmegen, The Netherlands. The dataset was collected and curated for research in computer aided diagnosis of prostate MR under supervision of dr. Huisman, Radboudumc as documented in: | Info |
|---|
| | G. Litjens, O. Debats, J. Barentsz, N. Karssemeijer and H. Huisman. "Computer-aided detection of prostate cancer in MRI", IEEE Transactions on Medical Imaging 2014;33:1083-1092. |
If you use this data for research then refer the above publication. The images come in two encodings. The acquired MR is provided in DICOM encoding. Additionally Ktrans images are provided. They come in mhd format. Ktrans is a key pharmacokinetic parameter computed from the available Dynamic contrast enhanced T1-weighted series. Each patient has one study with several DICOM images and one Ktrans image. The Ktrans image is encoded in two files ProstateX-[ProxID]-Ktrans.[mhd/zraw], where ProxID is the ProstateX patient identifier. The DICOM images comprise several Series each comprising several Instances. The DICOM files are documented in the ProstateX-Images.csv file. The columns in that file encode the following: ProxID – ProstateX patient identifier. Name – Series Description Studydate – Study Date fid – Finding ID Pos – Scanner Coordinate position of the finding WorldMatrix – Matrix describing image orientation and scaling ijk – image col,row,slice coordinate of finding ImageUID – Image Identifier TopLevel 0 - Series forms one image 1 – A set of Series forms a 4D image (e.g. Dynamic MR) NA – Series form one image, but is part of a Level 1 4D image
SpacingBetweenSlices – Scalar Spacing between slices VoxelSpacing – Vector with x,y,z spacing scalars Dim – Vector with 4D dimensions of the image DCMSerDescr – The original DICOM Series Description DCMSerUID – The DICOM Series UID DCMSerNum – The DICOM Series Number InstanceUIDList – DICOM Instances that make up this series ImageUIDList – TopLevel-NA Images the make up this Toplevel 1 image
For example, to get the ADC image of Patient ProstateX-0123 do the following. After you imported the DICOM files into your environment, go to patient ProstateX-0123 and find the series with ADC in it. In this case it is ‘ep2d_diff_tra_DYNDIST_ADC’. It has SeriesNumber 8. The DICOM images in that series form the ADC image for this challenge. Image slice j at coordinate i,j contains a finding fid. See findings for more details. FindingsThe findings are documented in the ProstateX-Findings.csv table. Documentation for the columns in that table is as follows: pos - Scanner Coordinate position of the finding ClinSig – Identifier available in training set that identifies whether this is a clinically significant finding. Either the biopsy GleasonScore was 7 or higher. Findings with a PIRADS score 2 were not biopsied and are not considered clinically significant. In our center the occurrence of clinically significant cancer in PIRADS 2 lesions is less than 5%.
|