- Created by natasha honomichl, last modified by Geri Blake on Mar 31, 2020
You are viewing an old version of this page. View the current version.
Compare with Current View Page History
« Previous Version 11 Next »
Summary
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- University of Texas M.D. Anderson Cancer Center, Houston, TX, USA - Special thanks to Kendall Kiser, MS Biomedical Informatics, from the Department of Radiation Oncology.
- The University of Texas Health Science Center School of Biomedical Informatics, Houston, TX, USA
Data Access
Click the Download button to save a ".tcia" manifest file to your computer, which you must open with the NBIA Data Retriever. Click the Search button to open our Data Portal, where you can browse the data collection and/or download a subset of its contents.
| Data Type | Download all or Query/Filter |
|---|---|
| Images (NIfTI, XX.X GB) | |
| Supplemental Data (CSV) |
|
Download buttons will not currently work until all data received and curated.
Click the Versions tab for more info about data releases.
Detailed Description
Image Statistics | |
|---|---|
Modalities | Seg |
Number of Patients | 402 |
Number of Studies | 402 |
Number of Series | |
Number of Images | |
| Images Size (GB) |
Table will be populated once all data received. Any additional data description will be added here.
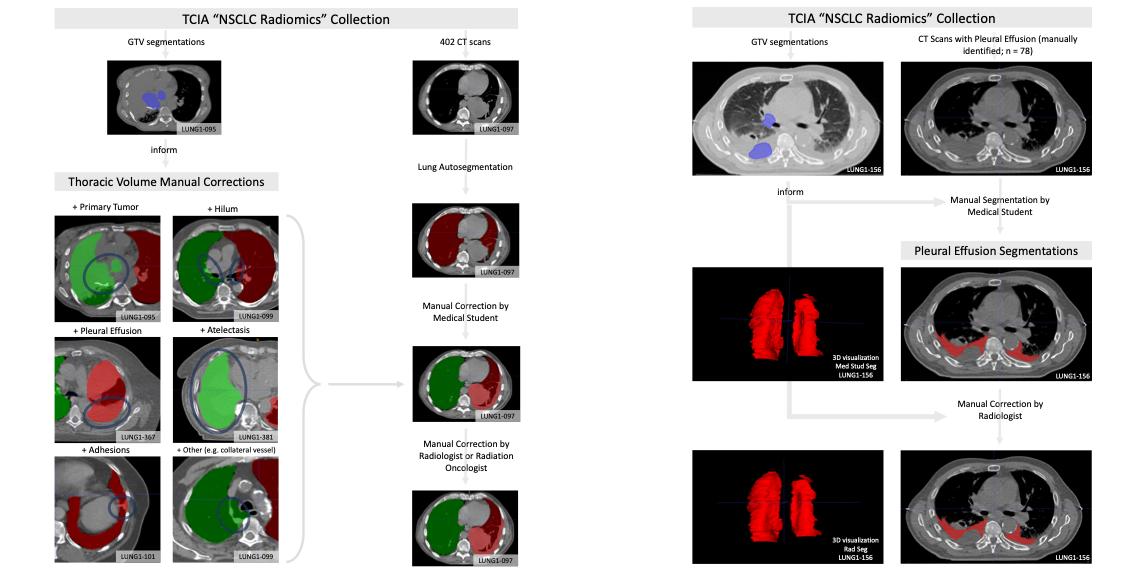
This dataset consists of left and right thoracic volume segmentations delineated on 402 CT scans from The Cancer Imaging Archive NSCLC Radiomics collection. Thoracic segmentations include lung parenchyma, tumor, atelectasis, adhesions, and effusion, when present. The initial thoracic segmentations were first generated automatically by a U-Net based algorithm trained on chest CTs without cancer, manually corrected by a medical student, and revised by a radiation oncologist or a radiologist.
On scans where effusion is present, separate segmentations labeling pleural effusion alone are also provided. Pleural effusion segmentations were manually delineated by a medical student and revised by a radiologist. Expert GTV segmentations already provided by the NSCLC Radiomics collection helped inform our segmentations, and areas of the effusion that overlap with GTVs are not included. Researchers interested in discriminating between GTV and effusion using imaging biomarker inputs may find our pleural effusion segmentations useful, especially when paired with the GTV segmentations provided in the NSCLC Radiomics collection.
Tabular data extending the tabular data accompanying the NSCLC Radiomics collection are also provided, including GTV, thorax, and effusion volumes (in cm3), tumor location, and image metadata. This dataset was utilized in a clinical informatics study correlating autosegmentation accuracy as gauged by several common metrics with time required for manual corrections.
Citations & Data Usage Policy
Add any special restrictions in here.
These collections are freely available to browse, download, and use for commercial, scientific and educational purposes as outlined in the Creative Commons Attribution 3.0 Unported License. Questions may be directed to help@cancerimagingarchive.net. Please be sure to acknowledge both this data set and TCIA in publications by including the following citations in your work:
Data Citation
Kiser, K.J., Ahmed, S. M. E. H., Stieb, S.M., Mohamed, A.S.R., Elhalawani, H., Park, P.Y.S., Doyle, N.S., Wang, B.J., Barman, A., Fuller, C.D., Giancardo, L. (2020). Data from the Thoracic volume and pleural effusion segmentations for symmetry-sensitive deep learning architecture development [Data set]. The Cancer Imaging Archive. https://doi.org/DOI goes here.
Acknowledgement
TCIA Citation
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: 10.1007/s10278-013-9622-7
Other Publications Using This Data
TCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk.
Version 1 (Current): Updated 2020/03/XX
| Data Type | Download all or Query/Filter |
|---|---|
| Images (NIfTI, xx.x GB) | (Requires NBIA Data Retriever.) |
| Supplemental Data (CSV) |
|
- No labels
