Summary
Magnetic Resonance Imaging (MRI) quality assessment measures were generated for- T1-weighted post-contrast axial MRI sequences from 133 The Cancer Genome Atlas Glioblastoma Multiforme Collection (TCGA-GBM) subjects
- T1-weighted post-contrast axial MRI sequences from 46 The Clinical Proteomic Tumor Analysis Consortium Glioblastoma Multiforme Collection (CPTAC-GBM) subjects
- Both T1- and T2-weighted axial MRI sequences from 54 The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma Collection (TCGA-CESC) subjects
All MRI scans for each cohort were downloaded as DICOM files from TCIA and then processed via MRQy to compute quality measures for (a) interrogating the presence of site- or equipment-specific variations within a cohort, and (b) quantifying the impact of MRI artifacts to determine what pre-analytical corrections are needed. The MRQy output can be easily interrogated via the associated HTML5 based front-end, allowing for real-time filtering and visualization. MRQy is available for download at: http://github.com/ccipd/MRQy. Manifest files to download the DICOM images these results were derived from are available on the Detailed Description page.
Acknowledgements
Research reported in this publication was supported by:
- National Cancer Institute of the National Institutes of Health under award numbers 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R01 CA216579-01A1, R01 CA220581-01A1, 1U01 CA239055-01, 1F31CA216935-01A1, NCI 1U01CA248226-01
- National Institute for Biomedical Imaging and Bioengineering (1R43EB028736-01)
- National Center for Research Resources under award number 1 C06 RR12463-01
- VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service
- The DoD Breast Cancer Research Program Breakthrough Level 1 Award (W81XWH-19-1-0668)
- The DOD Prostate Cancer Idea Development Award (W81XWH-15-1-0558)
- The DOD Lung Cancer Investigator-Initiated Translational Research Award (W81XWH-18-1-0440)
- The DOD Peer Reviewed Cancer Research Program (W81XWH-16-1-0329, W81XWH-18-1-0404)
- Dana Foundation David Mahoney Neuroimaging Program
- The V Foundation Translational Research Award
- The Ohio Third Frontier Technology Validation Fund
- The Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering
- The Clinical and Translational Science Award Program (CTSA) at Case Western Reserve University
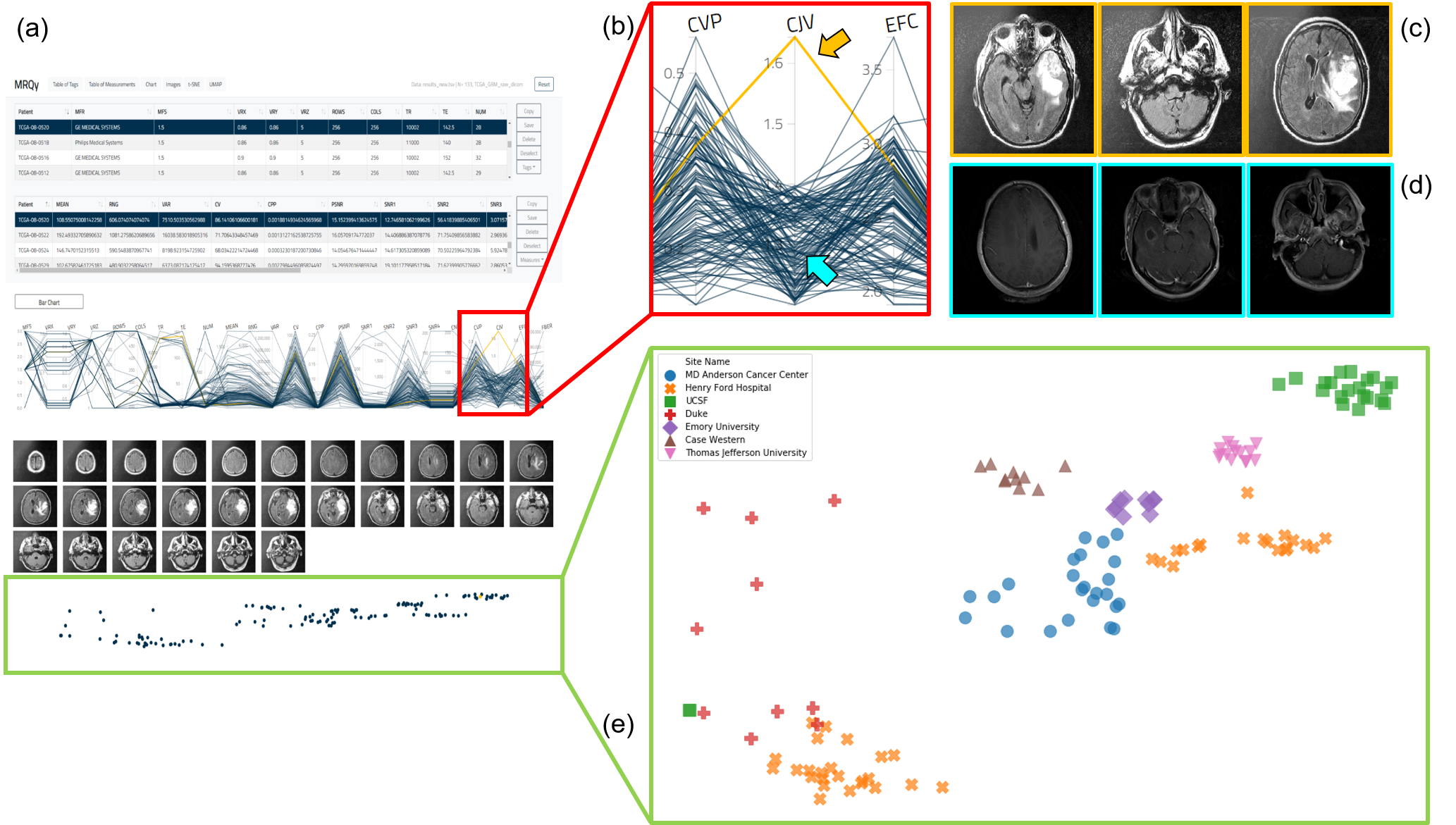
(a) MRQy front-end interface for interrogating TCGA-GBM cohort. (b) Outlier dataset identified on the parallel coordinate chart for the CJV quality measure found to exhibit shading artifacts on (c) representative images, especially when compared to (d) a different dataset without this artifact. (e) t-SNE scatter plot of quality measures revealing presence of site-specific batch effects (colors correspond to different sites, note presence of site-specific clusters).
Data Access
| Data Type | Download all or Query/Filter | License |
|---|---|---|
| TCGA-GBM MRQy results (49 kb, XLSX) | ||
| TCGA-CESC MRQy results (49 kb, XLSX) | ||
| CPTAC-GBM MRQy results (21 kb, XLSX) |
Click the Versions tab for more info about data releases.
Additional Resources for this Dataset
- Source code is available on GitHub at: http://github.com/ccipd/MRQy.
Collections Used in this Third Party Analysis
Below is a list of the Collections used in these analyses:
Source Data Type | Download | License |
|---|---|---|
Corresponding Original MR Images from TCGA-GBM (1.37GB) and CPTAC-GBM (DICOM, 0.4 GB) | (Download requires the NBIA Data Retriever and permission) | |
Corresponding Original MR Images from TCGA-CESC (DICOM, 2.72 GB) | (Download requires the NBIA Data Retriever) |
- The Cancer Genome Atlas Glioblastoma Multiforme Collection (TCGA-GBM)
- The Clinical Proteomic Tumor Analysis Consortium Glioblastoma Multiforme Collection (CPTAC-GBM)
- The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma Collection (TCGA-CESC)
Detailed Description
The spreadsheets provided are derived analyses from subsets of images from the The Cancer Genome Atlas Glioblastoma Multiforme Collection (TCGA-GBM), The Clinical Proteomic Tumor Analysis Consortium Glioblastoma Multiforme Collection (CPTAC-GBM), and The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma Collection (TCGA-CESC) collections. To download the original corresponding images for your own analysis please see the "Source Data Type" table.
Citations & Data Usage Policy
Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution should include references to the following citations:
Data Citation
Sadri, A. R., Janowczyk, A., Verma, R., Antunes, J., Madabhushi, A., Tiwari, P., & Viswanath, S. (2020). MRQy quality measures for TCIA MRI datasets [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/K9/TCIA.2020.JHZ2-T694
TCIA Citation
Clark, K., Vendt, B., Smith, K., Freymann, J., Kirby, J., Koppel, P., Moore, S., Phillips, S., Maffitt, D., Pringle, M., Tarbox, L., & Prior, F. (2013). The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. Journal of Digital Imaging, 26(6), 1045–1057. https://doi.org/10.1007/s10278-013-9622-7
Other Publications Using This Data
TCIA maintains a list of publications which leverage our data. If you have a manuscript you'd like to add please contact TCIA's Helpdesk.