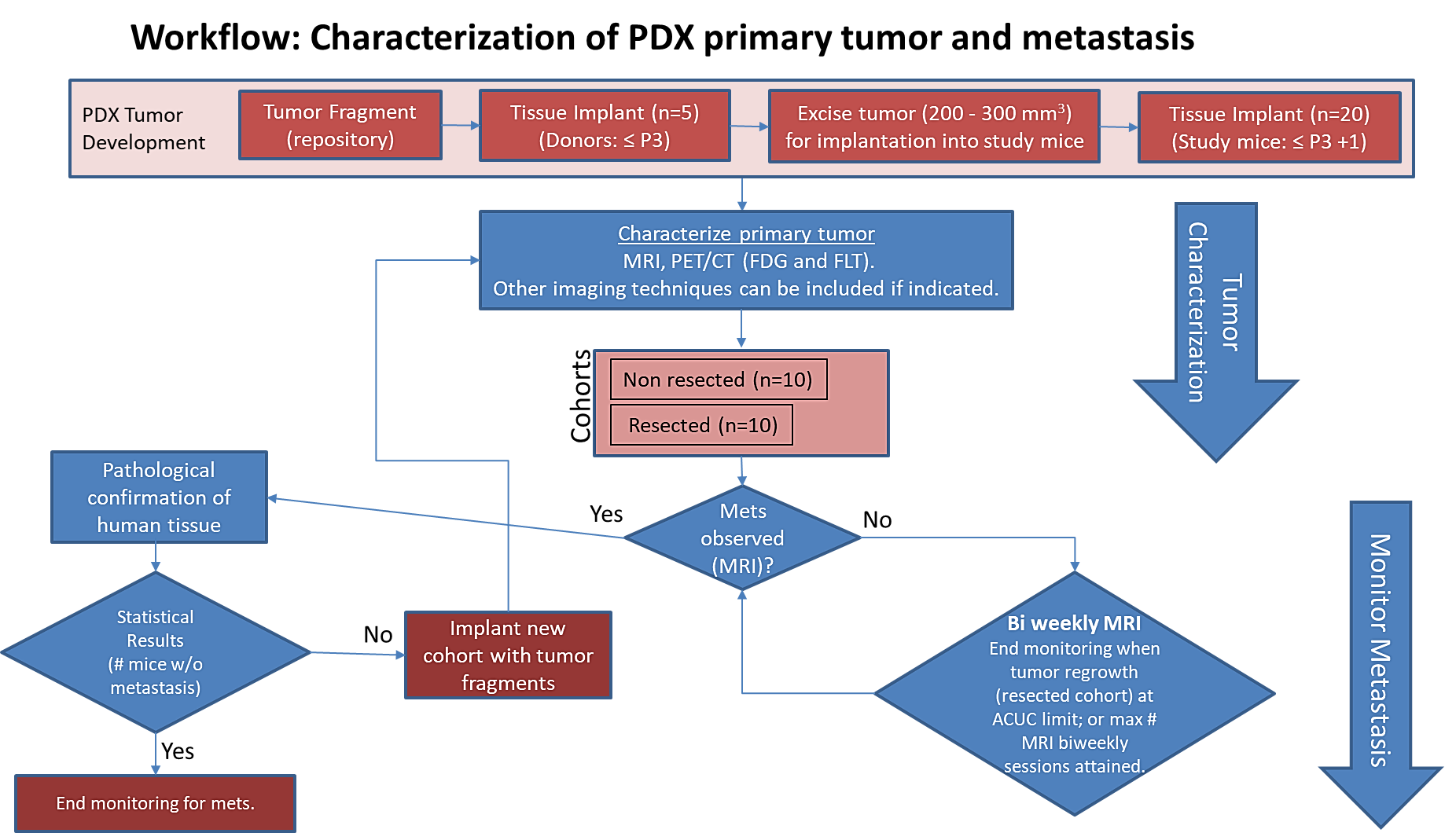
Summary
The imaging characteristics of this model (PDMR-425362-245-T) which is available from the National Cancer Institute Patient-Derived Models Repository (https://pdmr.cancer.gov/), is highly favorable for preclinical research studies of metastatic disease when used in conjunction with non-contrast T2 weighted MRI.
Results: Melanoma (PDMR-425362-245-T)
Table 1: Penetrance and location of pathological confirmed metastatic lesion(s).
# animals in group | # animals that displayed metastasis in MRI and confirmed by Pathology | Pathology confirmation of MRI (primary imaging site) | Other confirmed Location (s) | Mouse ID: MRI with pathology confirmation of metastasis |
10 (non-resected) | 4 (6 mice were EU due to xenograft size prior to observation of metastasis) | Lung | Kidney | 1512, 1516, 1518, 1520 |
10 (resected) | 10 | Lung | Kidney, Liver, Pancreas | 1506, 1508, 1509, 1510, 1511, 1515, 1517, 1519, 1521, 1523 |
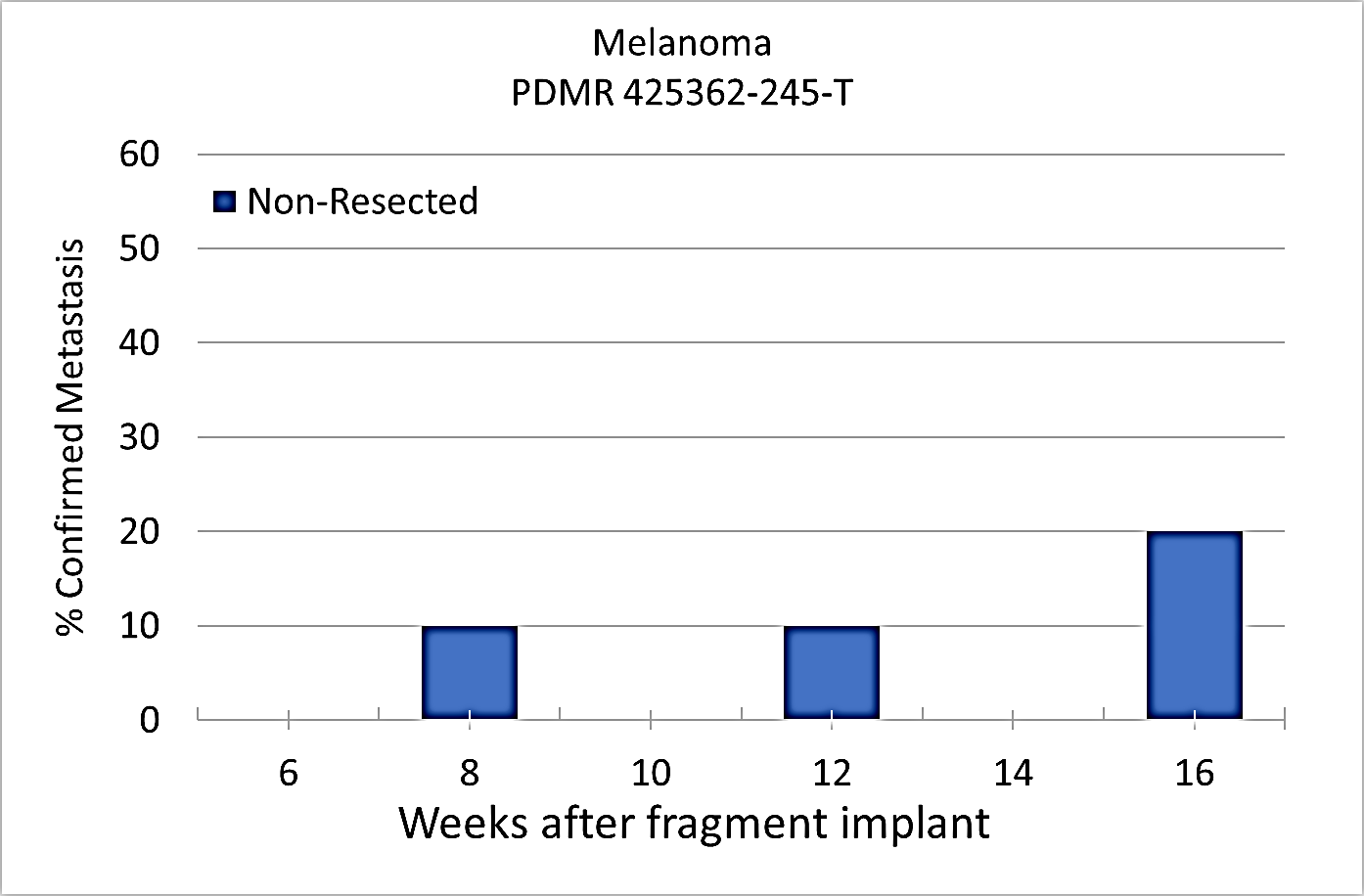
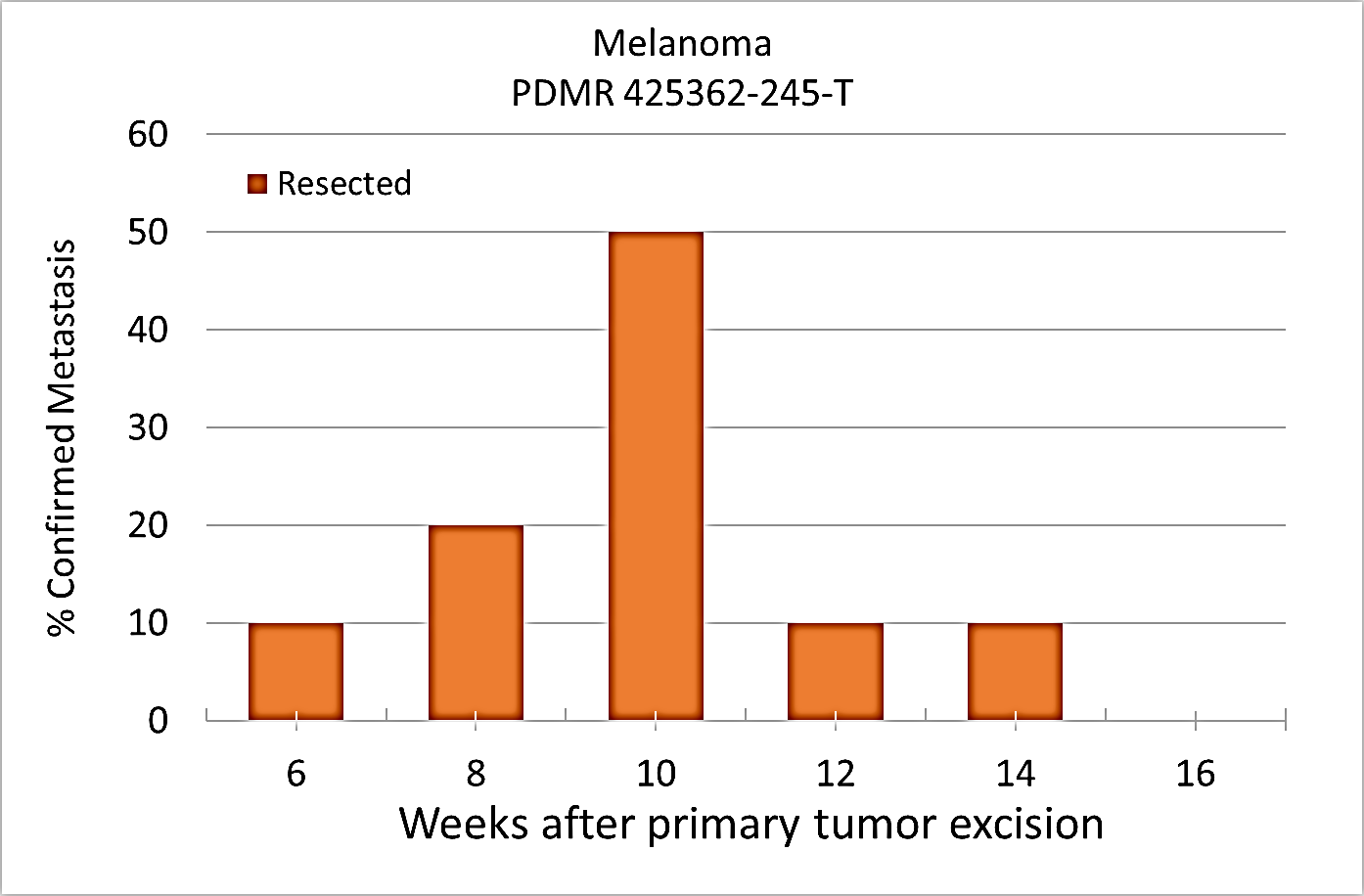
Percent penetrance with respect to the average time-to-metastasis for non-resected (plot A: time from implant) and resected (plot B: time from tumor resection) cohorts.
Plot A | Plot B |
|---|
PET/CT Characterization of the primary tumor: Baseline PET (SOP attached) were performed when tumor reached an approximate 200 mm3. Average SUVmax values (n=5) were calculated; [18F]FDG: 2.7 ± 0.5 and [18F]FLT: 2.0 ± 0.3.
Conclusion:
Melanoma PDMR-425362-245-T model can be challenging due to the rapid growth of the xenograft and regrowth. Metastases was well observed on T2 MRI imaging allowing non-invasive evaluation in treatment trials.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
- Frederick National Laboratory for Cancer Research – Special Thanks to Joseph D. Kalen, PhD, Lilia V. Ileva, MS, Lisa A Riffle, Nimit Patel, Keita Saito, PhD, Yvonne Evrard, PhD, Elijah Edmondson, DVM, PhD, Jessica Phillips, Simone Difilippantonio, PhD, Chelsea Sanders, Amy James, Lia Thang, Ulrike Wagner, Yanling Liu, PhD, John B. Freymann, and Justin Kirby.
- Division of Cancer Therapeutics and Diagnosis/National Cancer Institute - James L. Tatum, MD, Paula M Jacobs, PhD, Melinda G. Hollingshead, DVM, and James H. Doroshow, MD
- PixelMed Publishing – Special Thanks to David A. Clunie, MD
- University of Arkansas for Medical Sciences – Special Thanks to Kirk E. Smith
- This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract Number HHSN261201500003I. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Data Access
| Data Type | Download all or Query/Filter | License |
|---|---|---|
| Images (DICOM, 2.3 GB) | (Download requires NBIA Data Retriever) | |
| Tracking spreadsheet for PDMR-425362-245-T (XLSX, 40 kB) | ||
SR Conversion Routine (zip, 9 kB) (Converts a tracking spreadsheet into a DICOM Structured Report) | ||
| Standard Operating Procedure 50101 MRI T2 Weighted Non-Contrast Protocol Single Mouse Pulmonary Gated and Multi-Mouse Non-Gated (pdf, 133 kB) | ||
| Standard Operating Procedure 50102 Positron Emission Tomography (PET) imaging protocol (pdf, 140 kB) |
Click the Versions tab for more info about data releases.
Additional Resources for this Dataset
The National Cancer Institute (NCI) has developed a national repository of Patient-Derived Models (PDMs) comprised of patient-derived xenografts (PDXs), in vitro patient-derived tumor cell cultures (PDCs) and cancer associated fibroblasts (CAFs) as well as patient-derived organoids (PDOrg). These models serve as a resource for public-private partnerships and for academic drug discovery efforts. These PDMs are clinically-annotated with molecular information and made available in the Patient-Derived Model Repository. Data related to the specific subjects in this Collection can be found at:
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
Detailed Description
Image Statistics | |
|---|---|
Modalities | MR, SR |
Number of Subjects | 20 |
Number of Studies | 115 |
Number of Series | 210 |
Number of Images | 3509 |
| Images Size (GB) | 2.3 GB |
In addition to images, this collection includes Raw Data Storage SOP Class instances with MR Modality, generated by a Philips MR scanner; this data is not useful to anyone without the proprietary software to interpret it.
Citations & Data Usage Policy
Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution should include references to the following citations:
Data Citation
Tatum, J. L., Kalen, J. D., Jacobs, P. M., Ileva, L. V., Riffle, L. A., Keita, S., Patel, N., Sanders, C., James, A., Difilippantonio, S., Thang, L., Hollingshead, M. G., Phillips, J., Edmondson, E., Evrard, Y., Clunie, D. A., Liu, Y., Smith, K. E., Wagner, U., … Doroshow, J. H. (2020). Imaging characterization of a metastatic patient derived model of melanoma: (PDMR-425362-245-T) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.2020.7YRS-7J97
TCIA Citation
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: https://doi.org/10.1007/s10278-013-9622-7
Other Publications Using This Data
TCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk.
Version 2 (Current): Updated 2021/02/17
| Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM, 2.3 GB) | (Download requires NBIA Data Retriever) |
| Tracking spreadsheet for PDMR-425362-245-T | |
| SR Conversion Routine (Converts a tracking spreadsheet into a DICOM Structured Report) | |
| Standard Operating Procedure 50101 MRI T2 Weighted Non-Contrast Protocol Single Mouse Pulmonary Gated and Multi-Mouse Non-Gated | |
| Standard Operating Procedure 50102 Positron Emission Tomography (PET) imaging protocol |
Added two supplementary files: 1) an example tracking spreadsheet for multi-cohort murine studies and 2) an SR Conversion Routine to convert the tracking spreadsheet parameters into a DICOM Structured Report (SR).
Version 1: Updated 2020/05/22
| Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM, 2.3 GB) | (Download requires the NBIA Data Retriever) |
| Standard Operating Procedure 50101 MRI T2 Weighted Non-Contrast Protocol Single Mouse Pulmonary Gated and Multi-Mouse Non-Gated | |
| Standard Operating Procedure 50102 Positron Emission Tomography (PET) imaging protocol |