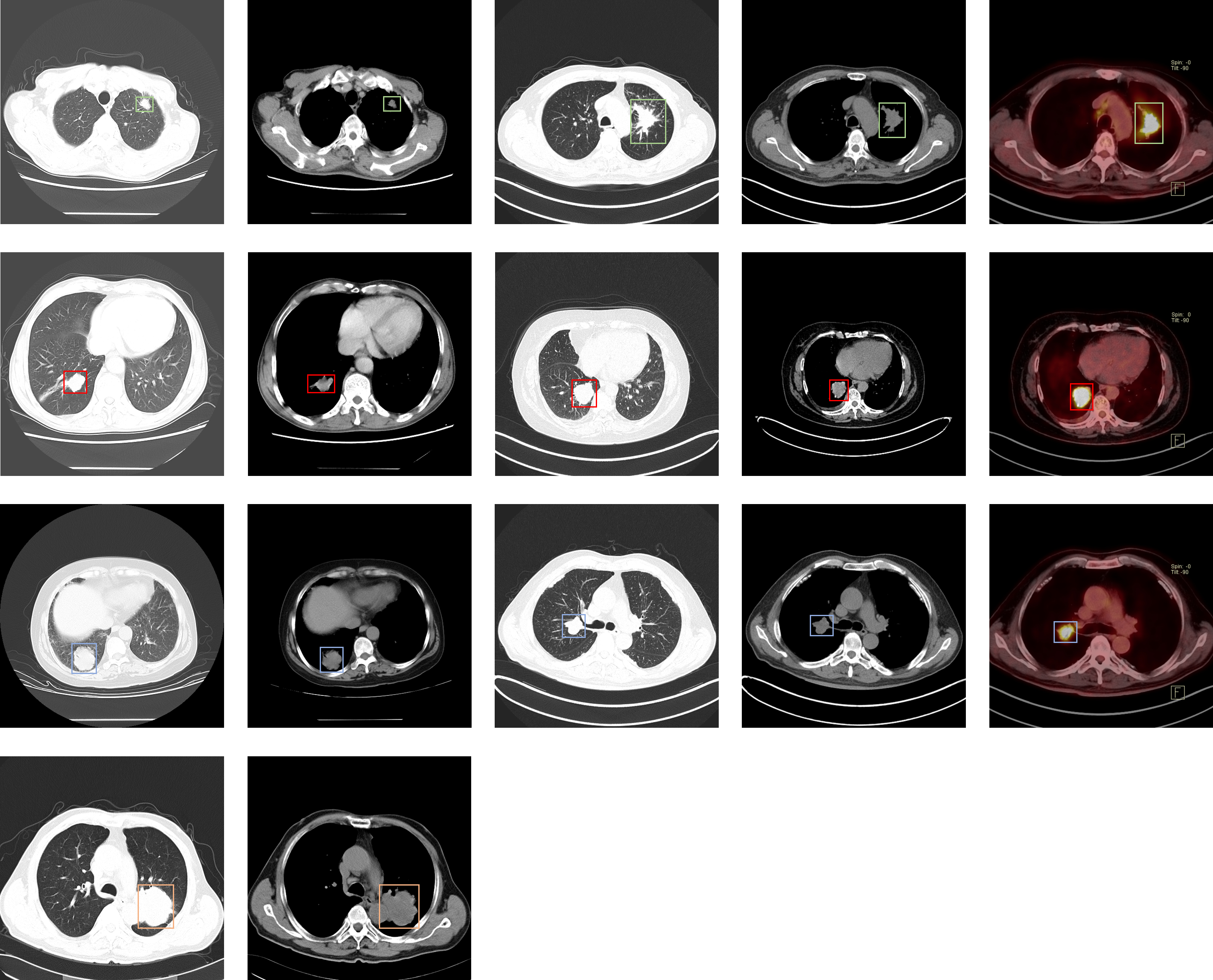
Summary
This dataset consists of CT and PET-CT DICOM images of lung cancer subjects with XML Annotation files that indicate tumor location with bounding boxes. The images were retrospectively acquired from patients with suspicion of lung cancer, and who underwent standard-of-care lung biopsy and PET/CT. Subjects were grouped according to a tissue histopathological diagnosis. Patients with Names/IDs containing the letter 'A' were diagnosed with Adenocarcinoma, 'B' with Small Cell Carcinoma, 'E' with Large Cell Carcinoma, and 'G' with Squamous Cell Carcinoma.
The images were analyzed on the mediastinum (window width, 350 HU; level, 40 HU) and lung (window width, 1,400 HU; level, –700 HU) settings. The reconstructions were made in 2mm-slice-thick and lung settings. The CT slice interval varies from 0.625 mm to 5 mm. Scanning mode includes plain, contrast and 3D reconstruction.
Before the examination, the patient underwent fasting for at least 6 hours, and the blood glucose of each patient was less than 11 mmol/L. Whole-body emission scans were acquired 60 minutes after the intravenous injection of 18F-FDG (4.44MBq/kg, 0.12mCi/kg), with patients in the supine position in the PET scanner. FDG doses and uptake times were 168.72-468.79MBq (295.8±64.8MBq) and 27-171min (70.4±24.9 minutes), respectively. 18F-FDG with a radiochemical purity of 95% was provided. Patients were allowed to breathe normally during PET and CT acquisitions. Attenuation correction of PET images was performed using CT data with the hybrid segmentation method. Attenuation corrections were performed using a CT protocol (180mAs,120kV,1.0pitch). Each study comprised one CT volume, one PET volume and fused PET and CT images: the CT resolution was 512 × 512 pixels at 1mm × 1mm, the PET resolution was 200 × 200 pixels at 4.07mm × 4.07mm, with a slice thickness and an interslice distance of 1mm. Both volumes were reconstructed with the same number of slices. Three-dimensional (3D) emission and transmission scanning were acquired from the base of the skull to mid femur. The PET images were reconstructed via the TrueX TOF method with a slice thickness of 1mm.
The location of each tumor was annotated by five academic thoracic radiologists with expertise in lung cancer to make this dataset a useful tool and resource for developing algorithms for medical diagnosis. Two of the radiologists had more than 15 years of experience and the others had more than 5 years of experience. After one of the radiologists labeled each subject the other four radiologists performed a verification, resulting in all five radiologists reviewing each annotation file in the dataset. Annotations were captured using Labellmg. The image annotations are saved as XML files in PASCAL VOC format, which can be parsed using the PASCAL Development Toolkit: https://pypi.org/project/pascal-voc-tools/. Python code to visualize the annotation boxes on top of the DICOM images can be downloaded here.
Two deep learning researchers used the images and the corresponding annotation files to train several well-known detection models which resulted in a maximum a posteriori probability (MAP) of around 0.87 on the validation set.
Acknowledgements
We would like to acknowledge the individuals and institutions that have provided data for this collection:
Drs. Huiping Han, Funing Yang and Rui Wang for their help collecting data
The Computer Center and Cancer Institute at the Second Affiliated Hospital of Harbin Medical University in Harbin, Heilongjiang Province, China for their help collecting the image data
Beijing Municipal Administration of Hospital Clinical Medicine Development of Special Funding (ZYLX201511)
Data Access
| Data Type | Download all or Query/Filter | License |
|---|---|---|
| Images (DICOM, 127.2 GB) | (Download requires the NBIA Data Retriever) | |
| Annotation Files (XML, 17.26 MB) | ||
| Clinical Data (XLSX, 36 KB) |
Click the Versions tab for more info about data releases.
Additional Resources for this Dataset
The NCI Cancer Research Data Commons (CRDC) provides access to additional data and a cloud-based data science infrastructure that connects data sets with analytics tools to allow users to share, integrate, analyze, and visualize cancer research data.
- Imaging Data Commons (IDC) (Imaging Data)
In addition, the following external resources have been made available by the data submitters. These are not hosted or supported by TCIA, but may be useful to researchers utilizing this collection.
- Annotations were captured using Labellmg
- The image annotations are saved as XML files in PASCAL VOC format, which can be parsed using the PASCAL Development Toolkit: https://pypi.org/project/pascal-voc-tools/
- Python code to visualize the annotation boxes on top of the DICOM images can be downloaded here.
Detailed Description
Image Statistics | Radiology Image Statistics |
|---|---|
Modalities | CT,PT |
Number of Participants | 355 |
Number of Studies | 436 |
Number of Series | 1,295 |
Number of Images | 251,135 |
| Images Size (GB) | 127.2 |
Citations & Data Usage Policy
Users must abide by the TCIA Data Usage Policy and Restrictions. Attribution should include references to the following citations:
Data Citation
Li, P., Wang, S., Li, T., Lu, J., HuangFu, Y., & Wang, D. (2020). A Large-Scale CT and PET/CT Dataset for Lung Cancer Diagnosis (Lung-PET-CT-Dx) [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/TCIA.2020.NNC2-0461
TCIA Citation
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. (2013) The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, 26(6):1045-1057. DOI: 10.1007/s10278-013-9622-7
TCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk.
Version 5 (Current): Updated 2020/12/22
Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM, 127.2GB) | (Download requires the NBIA Data Retriever) |
| Annotation Files (XML, 17.26 MB) | |
| Clinical Data (XLSX, 36 KB) |
Clinical data has been added for all 355 subjects.
Eight subjects were removed from the dataset because the submitting site determined that they required further medical examinations to make an accurate diagnosis.
Version 4: Updated 2020/10/16
Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM,132 GB) | (Download requires the NBIA Data Retriever) |
| Annotation Files (XML, 17.26 MB) |
Annotation files were corrected and updated at the request of the submitting site.
Version 3: Updated 2020/07/24
Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM,132 GB) | (Download requires the NBIA Data Retriever) |
| Annotation Files (XML, 14.62 MB) |
PET scans have been added for 140 subjects.
Version 2: Updated 2020/07/14
| Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM, 128 GB) | (Requires NBIA Data Retriever) |
| Annotation Files (XML, 14.62 MB) |
After publication of this dataset, the submitter notified us that the data for Subject Lung_Dx-A0266 really belonged to Subject Lung_Dx-A0251 and that Subject Lung_Dx-A0266 should not exist in the collection. Version 2 corrects this issue.
Version 1: Updated 2020/06/17
| Data Type | Download all or Query/Filter |
|---|---|
| Images (DICOM, 128 GB) | Unavailable, see version 2 note. |
| Annotation Files (XML, 14.62 MB) | Unavailable, see version 2 note. |