...
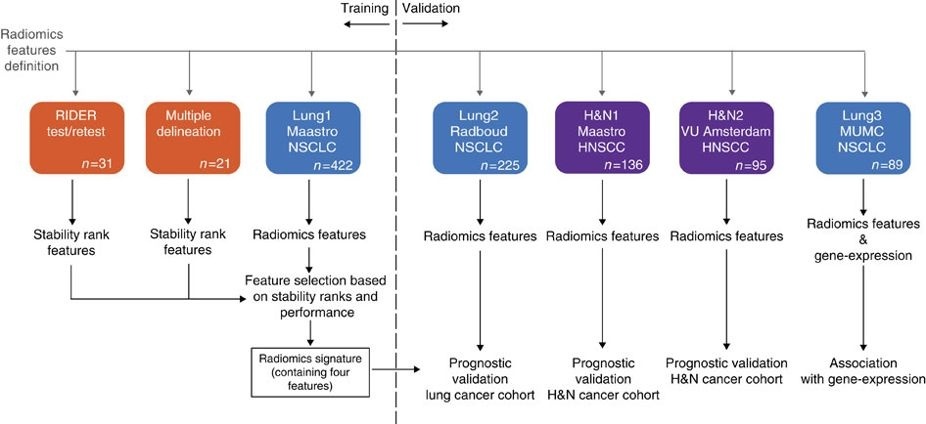
This data applies a radiomic approach to computed tomography data of 1,019 patients with lung or head-and-neck cancer which are described in Nature Communications (http://doi.org/10.1038/ncomms5006). The various arms of the study are represented in TCIA as distinct Collections including NSCLC-Radiomics (Lung1), NSCLC-Radiomics-Genomics (Lung3), Head-Neck-Radiomics-HN1 (H&N1), NSCLC-Radiomics-Interobserver1 (Multiple delineation), and RIDER Lung CT Segmentation Labels from: Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach (RIDER-LungCT-Seg) (RIDER test/retest).
Radiomics refers to the comprehensive quantification of tumour phenotypes by applying a large number of quantitative image features. In present analysis 440 features quantifying tumour image intensity, shape and texture, were extracted. We found that a large number of radiomic features have prognostic power in independent data sets, many of which were not identified as significant before. Radiogenomics analysis revealed that a prognostic radiomic signature, capturing intra-tumour heterogeneity, was associated with underlying gene-expression patterns. These data suggest that radiomics identifies a general prognostic phenotype existing in both lung and head-and-neck cancer. This may have a clinical impact as imaging is routinely used in clinical practice, providing an unprecedented opportunity to improve decision-support in cancer treatment at low cost.
...
| Localtab Group | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
...