| Redirect | ||||
|---|---|---|---|---|
|
Summary
| Excerpt |
|---|
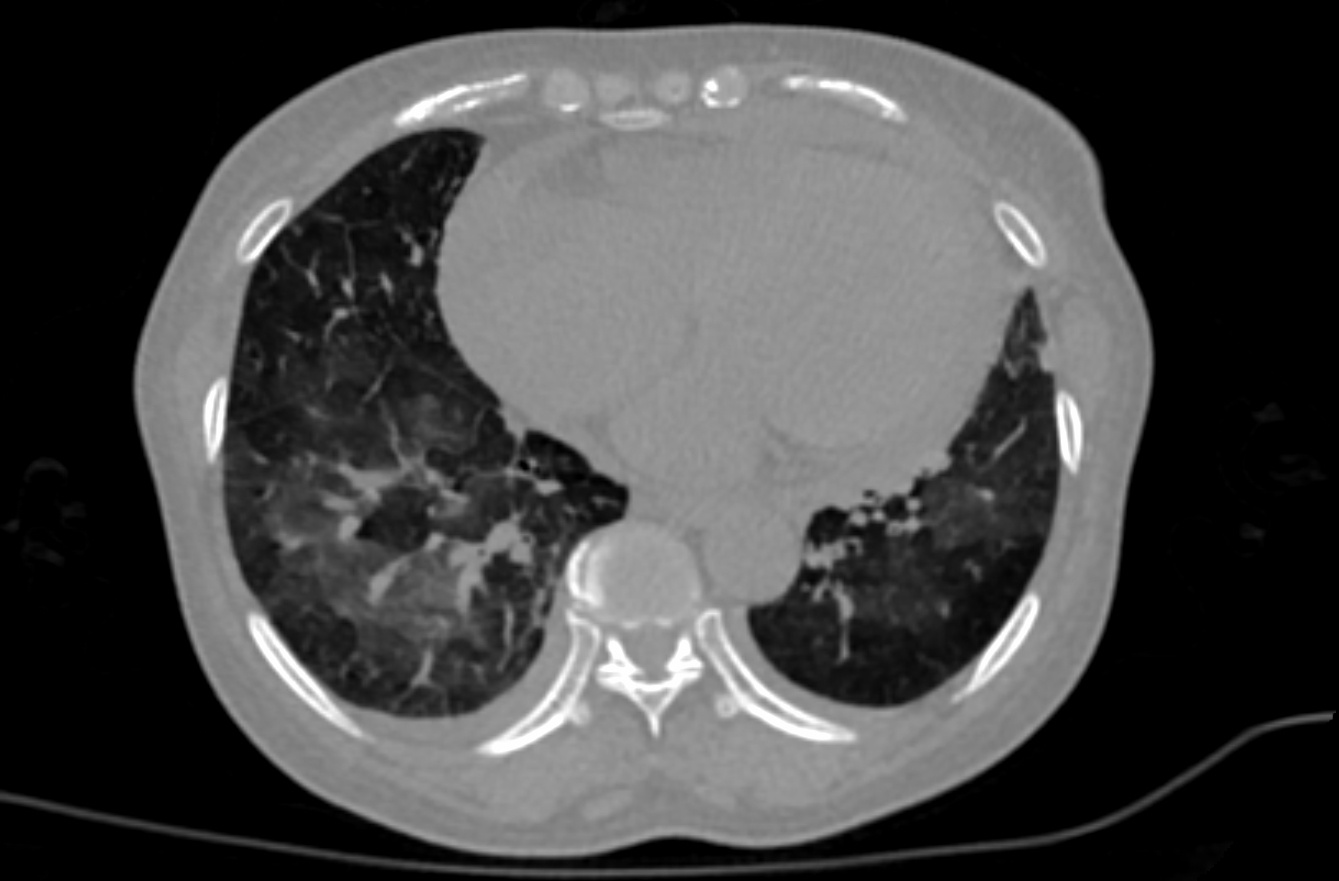
This dataset These retrospective NIfTI image datasets consists of unenhanced chest CT images of CTs:
The initial images for both datasets were acquired at the point of care in an outbreak setting from patients with Reverse in an outbreak setting, in NIfTI file format. The images were retrospectively acquired from a single region after CT of patients with Reverse Transcription Polymerase Chain Reaction (RT-PCR) confirmation for the presence of SARS-CoV-2. NIfTI CT images were converted from DICOM images. CTs were performed with intravenous contrast and a soft tissue reconstruction algorithm. Patients presented to a health care setting with a combination of symptoms, exposure to an infected patient, or travel history to an outbreak region. All patients had a positive RT-PCR for SARS-CoV-2 from a sample obtained within 1 day of the initial CT. This data may be a useful tool and resource for developing algorithms for medical applications in COVID-19, or data analysis challenges for the scientific communityCT exams were performed without intravenous contrast and with a soft tissue reconstruction algorithm. The DICOM images were subsequently converted into NIfTI format. The second dataset also had other follow up CTs, in addition to the initial point of care CT. A multidisciplinary team trained several models using portions of this data set (the first dataset, along with additional CTs and manually annotated images and other data &/or other CT’s)from other sources. A classification model derived in part from portions of this data (and also from other non-TCIA data) can be found the first dataset is described in a Nature Communications manuscript at: https://doi.org/10.1038/s41467-020-17971-2. Models partly derived from portions of this data (and also from other data not shown here), may be found at: The NVIDIA-related frameworks and models specific to this publication are available at no cost as part of the NVIDIA Clara Train SDK at https://ngc.nvidia.com/catalog/resourcescontainers/nvidia:clara:clara_ai_covid19_pipeline.A web-based research-only model (website for research use only) has CT drag-and-drop functionality. Upload of CT yields a return email that contains results. This model is also partly derived from portions of this data (and also from other non-TCIA data): https://marketplace.arterys.com/model/nvidiacovidCT. This COVID-19 classification model is detailed at: ai-covid-19. This includes both inference-based pipelines for evaluation, as well as model weights for further training or fine tuning in outside institutions. The second data set of 121 serial / sequential CTs in 29 patients is reported in a Scientific Reports manuscript at https://doi.org/10.1038/s41467s41598-020021-1797185694-25. |
Acknowledgements
The Imaging AI in COVID team would like to acknowledge the following individuals who supported this multi-disciplinary multi-national team effort:
- All frontline workers and Peng An, Sheng Xu, Evrim B. Turkbey, Stephanie A. Harmon, Thomas H. Sanford, Amel Amalou, Michael Kassin, Nicole Varble, Maxime Blain, Dilara Long, Dima Hammoud, Ashkan Malayeri, Elizabeth Jones, Holger Roth, Ziyue Xu, Dong Yang, Andriy Myronenko, Victoria Anderson, Mona Flores, Francesca Patella, Maurizio Cariati, Kaku Tamura, Hirofumi Obinata, Hitoshi Mori, Ulas Bagci, Daguang Xu, Hayet Amalou, Robert Suh, Gianpaolo Carrafiello, Baris Turkbey, Bradford J. Wood.
- Thanks for leadership support to: John Gallin, Steve Holland, Cliff Lane, Bruce Tromberg, Tom Misteli, Bill Dahut.
- Supported by the NIH Center for Interventional Oncology and the NIH Intramural Targeted Anti-COVID-19 (ITAC) Program.
TCIA COVID-19 Datasets
Additional datasets and information about TCIA efforts to support COVID-19 research can be found here.
| Localtab Group | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|