Summary
The objective of the ACRIN 6684 multi-center clinical trial was to determine the association of baseline FMISO PET uptake (maximal tumor to blood ratio, hypoxic volume) and MRI parameters (Ktrans, CBV) with overall survival, time to disease progression, and 6-month progression free survival in participants with newly diagnosed glioblastoma multiforme (GBM). The trial also collected standard brain cancer data such as Karnofsky performance status, but also pathological biomarkers that included MGMT status, HIF1-alpha, GLUT1, CAIX, CD31, and alpha-SMA expression assays. ACRIN 6684 Study Protocol After establishing eligibility and enrollment to the study, baseline imaging of both MR and PET was performed within 2 weeks of starting therapy. FMISO, has been helpful in evaluating tumor oxygenation status, which may affect how well it responds to radiation and chemotherapy. The MRI scans were designed to measure tumor characteristics related to oxygenation status, including changes in blood flow, blood volume, and blood vessel size. In the original protocol, following baseline imaging was an optional test-retest scan for FMISO PET only. Also included were PET and MRI scans at 3 weeks after the onset of chemo/radiation therapy, and 4 weeks following the end of standard treatment. Of the 50 patients enrolled in the study only one patient had a test-retest FMISO scan, and the requirement of scans mid and post therapy were dropped after the 4th case. The current protocol appears in Figure 1 below and can be found online (Protocol-ACRIN 6684 Amendment 7, 01.24.12) Figure 1. The latest protocol for ACRIN 6684 had PET and MR imaging performed only at baseline, up to 2 weeks prior to standard treatment (chemo + radiation therapy). Mid and post-therapy scans were eliminated from the protocol after Case 4, and only one patient had a retest FMISO scan. Of the 50 enrolled patients, 42 patients had evaluable imaging data for the primary aims of the study (see Gerstner et al. 2016). Acknowledgements This shared data set was provided by Elizabeth R. Gerstner, MD, James R. Fink, MD, David A. Mankoff, MD, PhD, and A. Gregory Sorenson, MD, in collaboration with the American College of Radiology Imaging network (ACRIN) with recognition to Alexander M. Spence, MD founding Co-PI for ACRIN 6684 who has since passed. Many thanks are due to the ACRIN 6684 trial team, and all the patients participating in the study. This study was supported by ACRIN, which received funding from the National Cancer Institute through UO1 CA080098, under the American Recovery and Reinvestment ACT of 2009 (ARRA) and UO1 CA079778.
Data Access
This is a limited access data set and is only available to members of NCI's Quantitative Imaging Network (QIN) until 8/31/2019. If you are a member of the QIN and would like to request access, please submit a CCP proposal to the QIN Coordinating Committee. Once access is granted, you can view and download these images on The Cancer Imaging Archive (TCIA) by logging in and selecting the ACRIN-FMISO-Brain collection.
Click the Versions tab for more info about data releases.
Detailed Description
Collection Statistics |
|
|---|---|
Modalities | MR |
Number of Patients | 45 |
Number of Studies | 370 |
Number of Series | 4626 |
Number of Images | 669,424 |
| Image Size (GB) | 95.6 |
Participant Eligibility and Enrollment:
Adult patients newly diagnosed with pathologically confirmed GBM (World Health Organization [WHO] grade IV) that had visible residual disease after surgical resection, and planned for initial treatment with radiation therapy (RT) and temozolomide (TMZ), with or without additional agents, were enrolled. Amount of residual tumor did not impact eligibility and visible residual disease included T2/FLAIR hyperintensity. The study enrolled the first patient in March 2010 and the last in August 2013, with follow up ending 1 year later (July 2014). Final accrual to the study was 50 patients, of which 42 were evaluable for the primary aim analysis as reported by Gerstner et al. 2016.
ACRIN 6684 MR Imaging
Of the 50 patients enrolled, 42 had evaluable imaging MR studies and 38 patients had evaluable 18F-FMISO PET scans relating to the primary aim. Additionally, 37 patients had evaluable DSC imaging, 31 had evaluable DCE imaging, 39 had evaluable diffusion tensor imaging (DTI) data, 17 had evaluable spectroscopy (MRS) data and 13 patients had BOLD imaging that has never been analyzed. For each MR imaging session, patient scans were completed on 1.5 or 3 T scanners (Philips 3T (12 patients), GE 3T (12 patients), Siemens 3T (2 patients), and Siemens 1.5T (five patients) magnets). The sequence of acquisition for each MR imaging session were as follows:
- Scout
- T1 Pre-contrast Spin Echo
- 3D T2 Rare
- FLAIR
- OPTIONAL: BOLD MRI 2D-EPI during transient inhalation of oxygen, minimum requirement 7 L/min using nasal cannula, 2 saturation levels were collected during this sequence.
- DCE (Dynamic contrast enhanced) 3D-T1 GRE imaging beginning with T1 mapping using 5 flip angles (2,10,15, 20, 30°). 1st injection of Gd.
- DWI/DTI (Diffusion weighted or diffusion tensor imaging) 2D EPI
- DSC (Dynamic susceptibility contrast) 2D EPI imaging following 2nd Gd Injection
- T1 Post-contrast 3D SPGR (MPRAGE)
- T1 Post-contrast Spin Echo
- MRS (magnetic resonance spectroscopy) 3D volumetric
The complete protocol for MR imaging can be found on the ACRIN website (MRI Parameters_062010.pdf).
ACRIN 6684 PET Imaging
Each FMISO PET imaging session consisted of acquiring a low-dose CT transmission scan used for attenuation correction and a PET emission scan at approximately 2 hours after injection of 18FMISO. During the emission scan, blood samples were acquired to determine the maximal tumor-to-blood ratio (TBmax) and the hypoxic volume (HV). Of the 42 patients with FMISO scans, 4 had no blood sampling performed. Patients were scanned using GE Discovery (31 patients), Siemens ECAT (6 patients) or Siemens Biograph (2 patients) PET scanners. Three patients (Cases 2, 3 and 5) were imaged on a Siemens prototype brain MR/PET scanner, that could not be calibrated to known activity units, such as Bq/cc.
Overview of Clinical Data
The basic data flow for legacy ACRIN multi-center clinical trials was that all clinical information provided by the local imaging sites were contained in a series of forms. The form data submitted by local investigators to ACRIN during and after the trial, were manually encoded into the ACRIN CTMS (Clinical Trial Management System), and were cross-checked for accuracy by ECOG-ACRIN personnel. These ACRIN 6684 forms (see the ACRIN 6684 Data Forms page), filled out by the local sites, deliver information on imaging, clinical management of the patient and pathology/outcome variables, like dates of progression and survival, along with other critical information. The image data was initially anonymized while uploading from the local sites through TRIAD software and archived in a DICOM database at ACRIN.
After the trial accrual had ended, the clinical data was sent to the Brown statistical center, that is funded by NCI to provide support for ECOG-ACRIN clinical trials, specifically for analysis of the primary and sometimes secondary aims of the trial. The statisticians at Brown strip all the actual dates, names and other PHI from the CTMS data and create a .csv file for each form that has selected information useful for analysis of the trial data. A Form Description file detailing all the forms used in the study accompanies the .csv data files. Additionally, the accompanying Data Dictionary file lists each element for each form that has been selected for data retention along with a description of each form element.
Extracting clinical (non-imaging) data example:
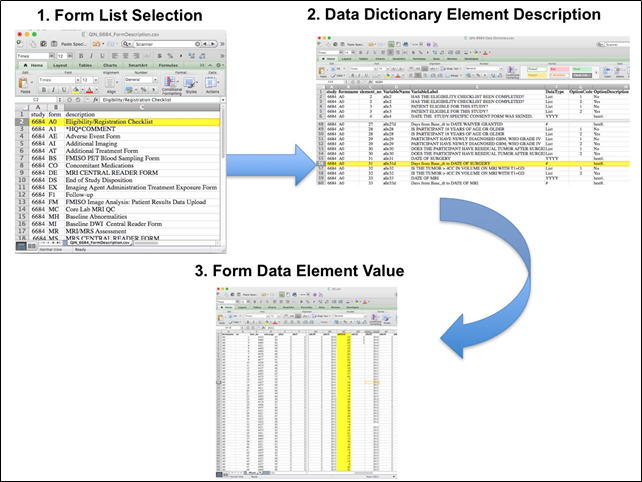
Beginning with the Form Description.csv file, select the form with the desired information needed, such as form A0.csv the patient Eligibility/Registration form. Next, using the Data Dictionary.csv file, find the form elements listed for A0 (eg., A0exx, where xx is the form element number). The file lists the form number, variable name, its description or label, the type of data, and, when applicable, the option codes and corresponding text values (option code:description pairs like 1=’No’, 2=’Yes’; or 1=’Baseline’, 2=’Post treatment’) for each data element available from the form. In the example in Figure 2, the A0 form element 31d (A0e31d) reports the days between the base date and the day of surgery for the patient. In the corresponding A0.csv file column G lists the days between the base date and surgery for each patient. ACRIN 6684 has about 30 forms and about 2000 form element descriptions listed in the Data Dictionary.
Figure 2: In this example of extracting clinical data, the first step is to 1) find the form from the form list, 2) Find the desired element and description in the Data Dictionary and finally 3) extract the values from the .csv data file.
All dates, like the visit date, are protected by presenting just the year; however, dates are also listed as offset days from the base date. The base date is typically the date the patient signs or agrees to participate in the clinical trial. The offset dates are used as a means of protecting patient information provided by the local sites in the original data, while allowing users to determine intervals between events. For example, the days between diagnosis and imaging might be determined by subtracting the days between the base date and imaging from the days between diagnosis and the base date.
The procedure above is basically how the statisticians organized the selected data for export, but the structure of the data dictionaries and individual forms are different for each clinical trial.
Citations & Data Usage Policy
This is a limited access data set and is only available to members of NCI's Quantitative Imaging Network (QIN) until 8/31/2019. If you are a member of the QIN and would like to request access, please submit a CCP proposal to the QIN Coordinating Committee. Upon receiving access you may only use it for the purposes outlined in your proposal. You are not allowed to redistribute the data or use it for other purposes. See TCIA's Data Usage Policies and Restrictions for additional details. Questions may be directed to help@cancerimagingarchive.net.
Please be sure to include the following citations in your work if you use this data set:
Data Citation
Kinahan, Paul; Muzi, Mark; Bialecki, Brian; Coombs, Laura. (2018). Data from ACRIN-FMISO-Brain. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA.2018.vohlekok
Publication Citation
Gerstner ER, Zhang Z, Fink JR, Muzi M, Hanna L, Greco E, Prah M, Schmainda KM, Mintz A, Kostakoglu L, Eikman EA, Ellingson BM, Ratai EM, Sorensen AG, Barboriak DP, Mankoff DA. ACRIN 6684: Assessment of Tumor Hypoxia in Newly Diagnosed Glioblastoma Using 18F-FMISO PET and MRI. Clin Cancer Res 22:5079-5086. 2016. PMC5065740. https://www.ncbi.nlm.nih.gov/pubmed/27185374. 10.1158/1078-0432.CCR-15-2529: 10.1158/1078-0432.CCR-15-2529.
Publication Citation
Ratai EM, Zhang Z, Fink J, Muzi M, Hanna L, Greco E, Richards T, Kim D, Andronesi OC, Mintz A, Kostakoglu L, Prah M, Ellingson B, Schmainda K, Sorensen G, Barboriak D, Mankoff D, Gerstner ER. ACRIN 6684: Multicenter, phase II assessment of tumor hypoxia in newly diagnosed glioblastoma using magnetic resonance spectroscopy. PloS one 13:1-14. 2018. PMC6002091.
TCIA Citation
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. (paper)
Other Publications Using This Data
TCIA maintains a list of publications which leverage our data. At this time we are not aware of any publications based on this data. If you have a publication you'd like to add please contact the TCIA Helpdesk.