- Created by natasha honomichl, last modified by Quasar Jarosz on Mar 30, 2020
You are viewing an old version of this page. View the current version.
Compare with Current View Page History
« Previous Version 16 Next »
Summary
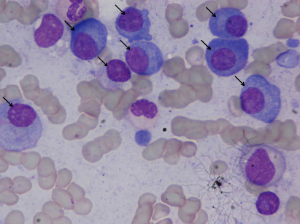 Microscopic images were captured from bone marrow aspirate slides of patients diagnosed with multiple myeloma as per the standard guidelines. Slides were stained using Jenner- Giemsa stain. Images were captured at 1000x magnication using Nikon Eclipse-200 microscope equipped with a digital camera. Images were captured in raw BMP format with a size of 2560x1920 pixels. In all, this dataset consists of 85 images. All these 85 images were stain normalized using our in-house methodology before being used for segmentation. These stain normalized images have been provided as the annotated dataset with plasma cells marked in all image slides contained in a presentation for the ready reference of readers.
Microscopic images were captured from bone marrow aspirate slides of patients diagnosed with multiple myeloma as per the standard guidelines. Slides were stained using Jenner- Giemsa stain. Images were captured at 1000x magnication using Nikon Eclipse-200 microscope equipped with a digital camera. Images were captured in raw BMP format with a size of 2560x1920 pixels. In all, this dataset consists of 85 images. All these 85 images were stain normalized using our in-house methodology before being used for segmentation. These stain normalized images have been provided as the annotated dataset with plasma cells marked in all image slides contained in a presentation for the ready reference of readers.Additional Notes
This collection has also been uploaded to the Harvard Blood Cancer Dataverse website. Please refer to DOI 10.7910/DVN/XCX7ST for more information.
Data Access
Click the Download button to browse and download the data from Box.
Click the Versions tab for more info about data releases.
Detailed Description
Image Statistics | |
|---|---|
Modalities | Pathology |
Number of Patients | 5 |
Number of Studies | 5 |
Number of Images | 85 |
| Images Size (GB) | 1.27 |
Citations & Data Usage Policy
These collections are freely available to browse, download, and use for commercial, scientific and educational purposes as outlined in the Creative Commons Attribution 3.0 Unported License. Questions may be directed to help@cancerimagingarchive.net. Please be sure to acknowledge both this data set and TCIA in publications by including the following citations in your work:
Data Citation
"Gupta, R., & Gupta, A. (2019). MiMM_SBILab Dataset: Microscopic Images of Multiple Myeloma [Data set]. The Cancer Imaging Archive. https://doi.org/10.7937/tcia.2019.pnn6aypl"
Publication Citation
[1] Anubha Gupta, Pramit Mallick, Ojaswa Sharma, Ritu Gupta, and Rahul Duggal, "PCSeg: Color Model Driven Probabilistic Multiphase Level Set based Tool for Plasma Cell Segmentation in Multiple Myeloma", Accepted, PLOSOne Journal, 2018.
[2] Anubha Gupta, Rahul Duggal, Ritu Gupta, Lalit Kumar, Nisarg Thakkar, and Devprakash Satpathy, “GCTI-SN: Geometry-Inspired Chemical and Tissue Invariant Stain Normalization of Microscopic Medical Images,”, under review.
[3] Ritu Gupta, Pramit Mallick, Rahul Duggal, Anubha Gupta, and Ojaswa Sharma, "Stain Color Normalization and Segmentation of Plasma Cells in Microscopic Images as a Prelude to Development of Computer Assisted Automated Disease Diagnostic Tool in Multiple Myeloma," 16th International Myeloma Workshop (IMW), India, March 2017.
TCIA Citation
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository, Journal of Digital Imaging, Volume 26, Number 6, December, 2013, pp 1045-1057. DOI: 10.1007/s10278-013-9622-7
Other Publications Using This Data
TCIA maintains a list of publications which leverage TCIA data. If you have a manuscript you'd like to add please contact the TCIA Helpdesk.
- No labels
