Please see the Data Usage Policies and Restrictions page for any questions about how our data can be used.
Questions about The Cancer Imaging Archive (TCIA) Website
1. Do you have a User's Guide?
A detailed TCIA user's guide can be obtained by clicking on TCIA User’s Guide.
2. How do I contact TCIA Help Desk?
The TCIA Help Desk can be reached in one of two ways:
- By email: help@cancerimagingarchive.net
- By phone or text message: +1 385-275-8242 (+1 385 ASK-TCIA)
3. What are the hours of operation for TCIA Help Desk?
The hours of operation are 7:00 am to 5:00 pm CST, Monday-Friday. Although the Help Desk is closed weekends and holidays, a request can be submitted at any time. Holiday hours and further information may be found on our Support page.
4. What if I have questions on navigating TCIA website?
The TCIA User Guide should provide all the direction you need. Please contact TCIA’s Help Desk with additional questions by calling or texting +1 385-275-8242 (+1 385 ASK-TCIA) or sending an email to help@cancerimagingarchive.net.
5. What if I am unable to connect to TCIA website?
Please contact TCIA’s Help Desk (+1 385-275-8242, voice or text ) or send us an email (help@cancerimagingarchive.net).
6. How do I submit feedback to TCIA website?
We welcome and appreciate your comments. Feedback maybe submitted via feedback@cancerimagingarchive.net.
7. What if I am being prompted to enter a verification code, but there is no verification code available and no place to type it?
A security warning prompt displays and asks "Do you want to view only the webpage content that was delivered securely". Answer NO to the prompt in order to see the captcha information that is used for verification.
If you answer YES, the captcha information will NOT display.
Questions about User Accounts
1. Do I need a user account to access TCIA?
Not unless you need to access private collections, or use shared lists. Just search for images without entering login information.
2. When I try to register after entering the captcha phrase, I get a PMW 5033 error or an error message stating, "The PWM configuration is invalid or corrupt. Please correct the error, or remove the configuration file." How do I proceed?
Assuming you are using Internet Explorer, change your browser's language to English by following these steps:
1. From the menu bar, select Tools.
2. Select Internet Options.
3. Click on General > Language and set to English (US).
3. How do I obtain a TCIA user ID and password?
From TCIA’s home page, click and then click New account. Remember that you do NOT need a user ID and password unless you need to access private collections. Just search for images without entering login information.
4. What if I have issues pertaining to my TCIA user ID or password?
Assuming that you already have an account, from TCIA’s home page, click and then click.
Choose the most appropriate of the following four options:
- You have an account but have forgotten your username.
- You have an account but have forgotten your password.
- You know your username and password, and would like to change your password.
- You would like to update your contact information in our system.
If you are still having trouble accessing your account, please contact the TCIA Support Center at help@cancerimagingarchive.net or call or text +1.385.275-8242 (+1.385.ASK-TCIA) during business hours for assistance.
Questions about Accessing Collections
1. How do I find additional information about a specific collection?
You can find a list of all collections on the TCIA home page: http://www.cancerimagingarchive.net. The middle window on this page is labeled "TCIA Collections". It contains a list of all collections in TCIA. The orange "Search TCIA" button takes you to the TCIA search page. Typing anything into the "Filter table" input box will restrict the list of collections to those which match the pattern in the box. To access detailed information about a collection, click the link in the "Collection" column. This will take you to a page in the wiki with detailed information about the collection.
You can reference the corresponding wiki pages for any particular collection when you are logged into TCIA. From the search page, in the collection area, mouse over and click on the question mark symbol for information on each collection. If there is a wiki available for the collection, click it.
2. How do I request access to a restricted collection?
To access a restricted collection, you can gain approval by calling or texting the Help Desk at +1 385-275-8242 (+1 385 ASK-TCIA), or send an email to help@cancerimagingarchive.net. Please indicate the name of the restricted collection.
3. How do I determine the status of a collection that has been submitted?
Please contact TCIA Help Desk (+1 385-275-8242, voice or text) or send an email (help@cancerimagingarchive.net) and indicate the name of the collection.
4. Is a listing of all of the modality abbreviations available?
Yes, modality abbreviations are listed here.
Questions about Searching for Images
1. How do I search and download images as well as find out information about a specific collection?
Using MRI images of the prostate as an example, follow the steps below to download images from a specific collection:
- Log into TCIA.
- To download all of the MRI prostate images, from the Simple Search filters, select MR under Image Modality and select prostate under Anatomical Site. The Summary tab shows updated pie charts and the number of image series in the search results.
- Click the Search Results tab.
- Click a Subject ID link to open all series associated with that subject.
Click the cart icon for one or more series. If you select only some of the series, the cart turns yellow. If you select all of the series, the cart turns green. The number of image series in your cart also appears next to the cart.
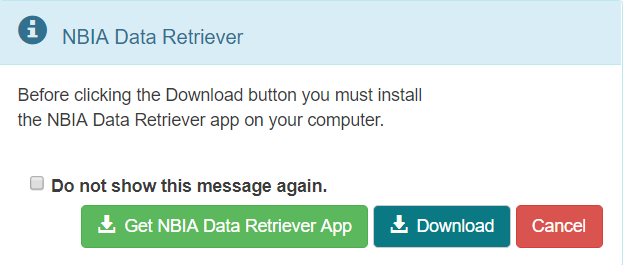
The cart itself, in the upper-right of the window, shows how many image series are in it and how large the download will be.Click .
A message appears reminding you to download the NBIA Data Retriever. If you have already downloaded it, click Do not show this message again and click . Otherwise, click to go to a page with links for downloads for each operating system.- Open the
manifest-xxx.tciafile in the NBIA Data Retriever. For more information on the NBIA Data Retriever, see NBIA Data Retriever FAQ. Also see question #1 in the previous section.
2. How do I search images?
The National Biomedical Imaging Archive (NBIA) software enables searching for images in TCIA’s local in vivo image repository. For simple searches, see Performing a Simple Search. For text searches, see Performing a Text Search (these links are in the TCIA User's Guide) for more detailed instructions.
3. What if my image search comes up empty?
If your query is too restrictive, the search will return no results. Try starting with one criterion and add additional criteria one at a time to refine your search.
4. How do I manage and view images in my cart?
As you review search results in TCIA, you can add items at any level to your cart for subsequent download. You can then view subject details, thumbnails of the image series, and DICOM headers.
5. Can I share the data in my cart?
Once you have added data to your cart, you can share it as a URL to a static set of data. When you or a colleague opens the URL in a browser window, the new TCIA Graphical User Interface (GUI) opens to the cart page.
6. What if I have a question pertaining to sharing a query?
You can save your query for later use or share it with a colleague by email.
7. How do I search for images showing a particular disease?
Collection descriptions provide information about what disease was studied. On the Search Images page, scroll down to the collections block and click the question mark to the left of the collection name. Click to see a brief description of the collection; it will usually have pointers to a site where more detailed information about the collection is available.
8. How do I search for images if I know the patient (subject) ID?
The NBIA software enables you to search for images in TCIA's image repository in a variety of ways. Although searching for a particular collection is the most common, you can also search for the images belonging to one or more subjects. Simply start on the Search page and enter one or more subject ID strings, separated by commas.
Questions about the NBIA Data Retriever
- Is NBIA Data Retriever open source?
- Do I have to use NBIA Data Retriever to pull data from the archive? Or are there alternatives?
- Where are the installation files for the current version of the NBIA Data Retriever?
- How do I know which version of the NBIA Data Retriever I am running?
- I generated a manifest file a while ago. How do I know which version of the NBIA Data Retriever to use with it?
- What are the dependencies of the NBIA Data Retriever on Linux and how we can satisfy the dependencies?
- How do I run the NBIA Data Retriever from the command line on RedHat/CentOS Linux?
- Failed to find library: jvm.dll
- How do I resolve the "xdg-desktop-menu: not found" error in the Ubuntu installation of the NBIA Data Retriever?
- How do resolve the “xdg-desktop-menu: No writable system menu directory found” error in the Ubuntu installation of the NBIA Data Retriever?
Is NBIA Data Retriever open source?
Yes! The NBIA Data Retriever, like the backend server it talks to, is licensed under the 3-clause BSD license, sometimes referred to as the 'new' or 'modified' BSD license. The BSD license is very permissive, business-friendly, and approved by the Open Source Initiative. The source code is available from GitHub in the CBIIT/NBIA-TCIA repository, specifically in the software/nbia-download-manager subdirectory. Developers are free to review or fork to create derivative works. Of course, we would appreciate, but do not require, pull requests back to the repository if you have created nice new features or bug fixes that others might benefit from. They would be reviewed by the NBIA development team for inclusion, possibly after some tweaks, back into the main repository.
If you do create a derivative work, please keep in mind that the RESTful API that the NBIA Data Retriever uses to communicate back to the NBIA services is not standardized. The API could change in future versions of NBIA or the NBIA Data Retriever, though we do try to maintain some level of backwards compatibility. The same is true of the format for the manifest files that drive the NBIA Data Retriever. It is up to the maintainer of derivative works to stay up-to-date on the RESTful API and manifest file format.
Do I have to use NBIA Data Retriever to pull data from the archive? Or are there alternatives?
Data in the archive is available through other methods. For example, a person versed in scripting or programming could easily pull data from the archive using the TCIA or NBIA Search RESTful interfaces, described at TCIA Programmatic Interface REST API Guides. Other organizations and people have also created software that can access data in TCIA. Many, but not all, of these tools and sites are listed in the Data Analysis Centers section of TCIA's web site. If you are interested in using TCIA data in the cloud, please check out NCI's Imaging Data Commons, which hosts public subsets (not all – yet) of TCIA data on a cloud platform for use by researchers. (IDC is still growing, and has not yet reached its full plan yet.)
Where are the installation files for the current version of the NBIA Data Retriever?
- RPM (tested on centOS)
To run this file, type the following at the command prompt:sudo yum -v -y remove NBIADataRetriever-4.4.x86_64.rpm;sudo yum -y install NBIADataRetriever-4.4.1.x86_64.rpm - DEB (tested on Ubuntu)
To run this file, type the following at the command prompt:sudo -S dpkg -r nbia-data-retriever-4.4.deb;sudo -S dpkg -i nbia-data-retriever-4.4.1.deb - NBIA Data Retriever in the Mac App Store
- Windows
How do I know which version of the NBIA Data Retriever I am running?
For NBIA Data Retriever version 3.0 and later, the version information appears in the About menu. The version also appears at the bottom of the login windows when you download a private collection. The following steps explain how to find out which version of the app you are running for version 2.0 and earlier in different operating systems.
On Windows 10:
- Open the Start menu.
- Click Settings > System.
- From the left pane, select Apps & features.
- Search for NBIA Data Retriever.
- Click NBIA Data Retriever and the version number appears in the application details.
On Mac:
- Go to the Finder.
- Click an application to select it, then press the spacebar. A window appears, showing the version number, size, and last modification date.
On CentOS:
- Type the following command at the command prompt of a terminal window:
yum list installed | grep "NBIA"
On Ubuntu:
- Type the following command at the command prompt of a terminal window:
dpkg -l |grep nbia
I generated a manifest file a while ago. How do I know which version of the NBIA Data Retriever to use with it?
Always use the latest version of the application. The application is backward-compatible. The following table shows the version compatibility between manifest files and the NBIA Data Retriever.
NBIA Data Retriever Version | Manifest v1.0 | Manifest v2.0 | Manifest v3.0 |
|---|---|---|---|
1.0 | Working | Working with public data requests* | Not working |
2.0 | Working with public data requests | Working | Auto Upgrade and Manual Upgrade are working. Remind Me Later does nothing. |
3.0 | Working with public data requests | Working | Working |
| 4.0 | Working with public data requests | Working | Working |
| 4.1 | Working with public data requests | Working | Working |
What are the dependencies of the NBIA Data Retriever on Linux and how we can satisfy the dependencies?
On CentOS or Red Hat Linux, use the command sudo yum -v -y remove NBIADataRetriever-4.3-1.x86_64.rpm;sudo yum -y install NBIADataRetriever-4.4-1.x86_64.rpm to install the NBIA Data Retriever. This command pulls and installs all missing dependent libraries when the NBIA Data Retriever is installed. If you install another way, you may not install all of the dependent libraries. Also, note that the NBIA Data Retriever always takes a manifest file as a parameter. In a GUI desktop environment, start the NBIA Data Retriever by clicking a manifest file.
How do I run the NBIA Data Retriever from the command line on RedHat/CentOS Linux?
The NBIA Data Retriever has two variants for Linux: the Graphical User Interface (GUI) app and a command-line interface. The command-line interface is available for the NBIA Data Retriever starting with version 4.1 and documentation is available at NBIA Data Retriever Command-Line Interface Guide.
The GUI app can also be started on the command line but needs the support of a desktop environment. For NBIA Data Retriever 3.2 and later, you can start the NBIA Data Retriever this way by entering the following command at the command prompt:
sudo /opt/NBIADataRetriever/NBIADataRetriever <full path of manifest file including the file name>
This command assumes that the NBIA Data Retriever is installed on /opt/Downloader, which is the location it is installed at when installed with the command sudo yum -v -y remove NBIADataRetriever-4.3-1.x86_64.rpm;sudo yum -y install NBIADataRetriever-4.4-1.x86_64.rpm.
Once the NBIA Data Retriever is installed on the desktop, you can click any manifest file with the extension .tcia to invoke the app.
The NBIA Data Retriever requires a manifest file to run. You can create a manifest file in the TCIA Radiology Portal and with an API.
Failed to find library: jvm.dll
I clicked a manifest file and received the error “Failed to find library: jvm.dll” on Windows 10. How can I fix it?
This is a Java bug. This issue has a workaround. You must uninstall the NBIA Data Retriever and download the free community version of Visual Studio 2017. Install Visual Studio 2017 and then reinstall the NBIA Data Retriever.
How do I resolve the "xdg-desktop-menu: not found" error in the Ubuntu installation of the NBIA Data Retriever?
If the Ubuntu server does not have a desktop environment installed, it will not have the “xdg-utils” package. To solve this problem, run the following command.
$cd /tmp $wget https://cbiit-download.nci.nih.gov/nbia/releases/ForTCIA/NBIADataRetriever_4.1/nbia-data-retriever-4.1.deb $sudo apt install xdg-utils $sudo -S dpkg -r nbia-data-retriever-4.0.1.deb;sudo -S dpkg -i nbia-data-retriever-4.1.deb
How do resolve the “xdg-desktop-menu: No writable system menu directory found” error in the Ubuntu installation of the NBIA Data Retriever?
Ubuntu has a bug that results in this error. To solve this problem, run the following command, then run the installation command again.
sudo mkdir /usr/share/desktop-directories/
Questions about Downloading Images
1. How do I download images in JPEG, TIFF, or BMP rather than DICOM format?
You can download items you add during the current work session. Before you download items you must download and then install the NBIA Data Retriever.
You select items to download by first searching for them in TCIA and then adding them to your cart.
The legacy Download Manager permits downloading of DICOM images only. To capture a large number of images, download them in DICOM and then reference the websites below for assistance with utilities that will convert DICOM images to other formats.
To download only a single image (or a handful of images one at a time):
- In the NBIA application, choose a study and series, then Show Images;
- Right-click over the image and Save Picture As .(jpeg).
2. How do I download annotated image files?
The .XML files are in the series folder since they are related to the images in that series. You can find the annotated files by viewing the list of files in a given folder.
3. What Java version is required for Download Manager?
Java version 6 or higher of the Java Runtime Environment (JRE) must be installed to be able to open the Download Manager. See Download Manager for more information.
4. What if I receive an error message from Download Manager or Download Manager hangs?
The number of series that can be downloaded using Download Manager is dependent on several factors: average image size, RAM on your computer, free disk space on your computer, network speed, and Java limitations. If you experience issues or errors associated with the download, reduce the number of series in your basket and try again.
If you never even see the Download Manager GUI, and Java reports that the Download Manager failed to launch, you may be running an old version of Java, and/or your Java application cache may be stale and needs to be cleared. See Clearing the Java Application Cache for instructions on how to clear the cache.
In some cases, the first attempt to run Download Manager on a computer might fail. Please simple retry launching the download manager (i.e., click on the "Download Manager" link on the shopping cart page). If the error persists, please contact TCIA's helpdesk at help@cancerimagingarchive.net , letting them know what computer, operating system version of Java that you are using. You can find instructions for determining this information at this link (click here).
5. I am behind a proxy server, will Download Manager work?
Accessing TCIA’s Download Manager directly through a proxy server is currently not supported. If your only access to the Internet is through a proxy server, our current recommendation is to ask your IT Department to allow your workstation to bypass the proxy server for direct access to https://public.cancerimagingarchive.net/ncia.
Java may be configured to utilize proxy settings; however, the developers of NBIA have not tested whether Download Manager correctly handles such settings.
6. Does Download Manager work with any browser?
Theoretically, yes. However, we principally test with Firefox, Chrome, and Internet Explorer. We also test with Safari (the default browser on most Macs), and it did work up to version delivered on Mac OS X version 10.8.3 (Mountain Lion). After 10.8.3, Safari will silently download the JNLP file used to launch the Download Manager, but will not automatically start the Download Manager program. Safari does not give the user any warning, so to the casual user it appears that the Download Manager link is broken. One has to go to the Downloads folder, find the JNLP file just downloaded, and then command | control | right click the JNLP file and select "Run" from the pop-up context menu to start the Download Manager.
Some browsers on some operating systems do not have our Certificate Authority in their list of trusted root Certificate Authorities. In these cases, the browser may present a warning about not trusting the connection to the web site. You may still be able to download images by clicking on the "Continue" or similar button.
7. Are there operating system requirements for Download Manager?
In theory, the Download Manager, being Java based, will work on any operating system. However, the Mac OS X operating system has been imposing additional security requirements on Java Web Start programs, such as the Download Manager, which make it difficult to run them without having administrative access to the Mac. In particular, the Gatekeeper function in 10.8.4 (Mountain Lion) and later versions of Mac OS X by default blocks starting third party programs that have not been either obtained from the Apple App store, or that have not been signed with an Apple Developer ID. This includes, naturally, Java Web Start programs such as the Download Manager. To get around this restriction, one can either change the default Gatekeeper setting, found in the Security tab of Apple System Preferences, to allow the running of 3rd party applications, or download the JNLP file used to launch the Download Manager, and then command | control | right click the JNLP file and select "Run" from the pop-up context menu. Note that either method requires either having administrative access to the Mac OS, or knowing the username/password of an administrator account, since the Mac will prompt for this information.
8. What if I am unable to download images through Download Manager and receive an error message in the status bar?
Our support team needs the following information to perform additional troubleshooting:
1. A screen capture of the error message.
2. A screen capture of the Java console (you may need to enable viewing the Java console using the Control Panel > Java Control Panel > Advanced > Settings: Java Console > Show Console).
3. Operating system and version.
- Windows XP: Start > Control Panel > System > General tab.
- Windows 7: Start > Control Panel > System and Security > System.
4. Browser and version.
- Internet Explorer: From the menu bar, select Help > About Internet Explorer.
9. How many series can Download Manager download from a single shopping cart?
Theoretically, there is no maximum number of series that Java allows Download Manager to hold in a single shopping cart. However, it is not practical to download more than around 3500 series at one time. The number of series in your basket will display on the left navigator at the top under My Data Basket > View Contents.
10. Where does Download Manager save my images?
When downloading images the default destination location for Windows devices is a path that is hidden by default which makes it hard to locate after the download completes. We suggest clicking the "Browse" button and selecting a more convenient destination for your data.
Questions about the TCGA-related imaging data
How do I find additional information about brain tumors?
Please refer to The Cancer Genome Atlas-Glioblastoma Multiforme (TCGA-GBM) collection for well-documented brain-tumor images as described here: https://wiki.cancerimagingarchive.net/display/Public/TCGA-GBM. (This wiki page also links to pages describing related research activities and related resources.)
In the Related Data section, there is a link to a blank GBM Case Quality Control Form that details the kinds and descriptions of clinical data collected. The data are freely available from the TCGA Data Portal: https://tcga-data.nci.nih.gov/tcga.
Once there, click on the link to GBM data. Since TCIA does not host the TCGA Data Portal, we do not offer guidance for browsing the Data Portal.
The TCGA-Lower Grade Glioma (LGG) collection is another set of brain tumor images: TCGA-LGG.
Questions about Tumors
Do the exams in TCIA show both malignant and benign tumors?
All patients who are part of TCIA at this time are cancer-positive by definition. We have no collections of non-malignant tumors or healthy patients. The only exception would be patients with multiple tumors who might have a benign tumor in addition to a malignant tumor, but at least one tumor will be malignant in each patient.
Questions about Limited Access Collections
How do I access National Lung Screening Trial (NLST) data?
Access to NLST images and data requires approval of the National Cancer Institute (NCI). For details, please see: https://biometry.nci.nih.gov/cdas/studies/nlst.
Useful References
MRI Basic Science and Abbreviations
Study materials for physicists and other clinical scientists learning the basics of magnetic resonance imaging (MRI): http://www.revisemri.com. Abbreviations only: http://www.revisemri.com/questions/misc/mri_abbrev
The site gives me problems, and error windows have popped up while I am trying to search or download images.
Our technical support team may ask you to answer these three questions if you're having difficulty with our website:
Which operating system are you using? (Typical examples would be Windows8, Mac OSX, Redhat Linux)
- Which browser (and version of that browser) are you using? (Typical examples would be Safari, Internet Explorer, Firefox, Chrome)
- Is Java installed on your machine, (and which version of that java are you using)? (Typical example would be yes, Java7 (u67))
To find your OS, browser, and java version, you may find the guide at Discover your OS Browser and Java Version helpful. The next step would be to test the site (but not necessarily click "download my basket") on another machine like a home laptop, to see if perhaps your institution has set, for example, a special layer of security on its network. The next step would be to email help@cancerimagingarchive.net to schedule an appointment for a webex teleconference with our technical support team to work with you interactively to facilitate access.